-
Notifications
You must be signed in to change notification settings - Fork 15
Linking it all together
John Colby edited this page Apr 15, 2011
·
18 revisions
The individual processing steps that we saw in Basic Workflow can be linked together into a core pipeline for along tract processing.
trkPath = fullfile(subDir, 'CST_L.trk');
volPath = fullfile(subDir, 'dti_fa.nii.gz');
[header tracks] = trk_read(trkPath);
volume = read_avw(volPath);
tracks_interp = trk_interp(tracks, 100);
tracks_interp = trk_flip(header, tracks_interp, [97 110 4]);
tracks_interp_str = trk_restruc(tracks_interp);
[header_sc tracks_sc] = trk_add_sc(header, tracks_interp_str, volume, 'FA');
[scalar_mean scalar_sd] = trk_mean_sc(header_sc, tracks_sc);We can time these steps to get a feel for how long it might take if we were to scale up the process over many tracts and subjects.
profile on
tic
trkPath = fullfile(subDir, 'CST_L.trk');
volPath = fullfile(subDir, 'dti_fa.nii.gz');
[header tracks] = trk_read(trkPath);
volume = read_avw(volPath);
tracks_interp = trk_interp(tracks, 100);
tracks_interp = trk_flip(header, tracks_interp, [97 110 4]);
tracks_interp_str = trk_restruc(tracks_interp);
[header_sc tracks_sc] = trk_add_sc(header, tracks_interp_str, volume, 'FA');
[scalar_mean scalar_sd] = trk_mean_sc(header_sc, tracks_sc);
toc
profile viewerOutput:
Elapsed time is 4.013543 seconds.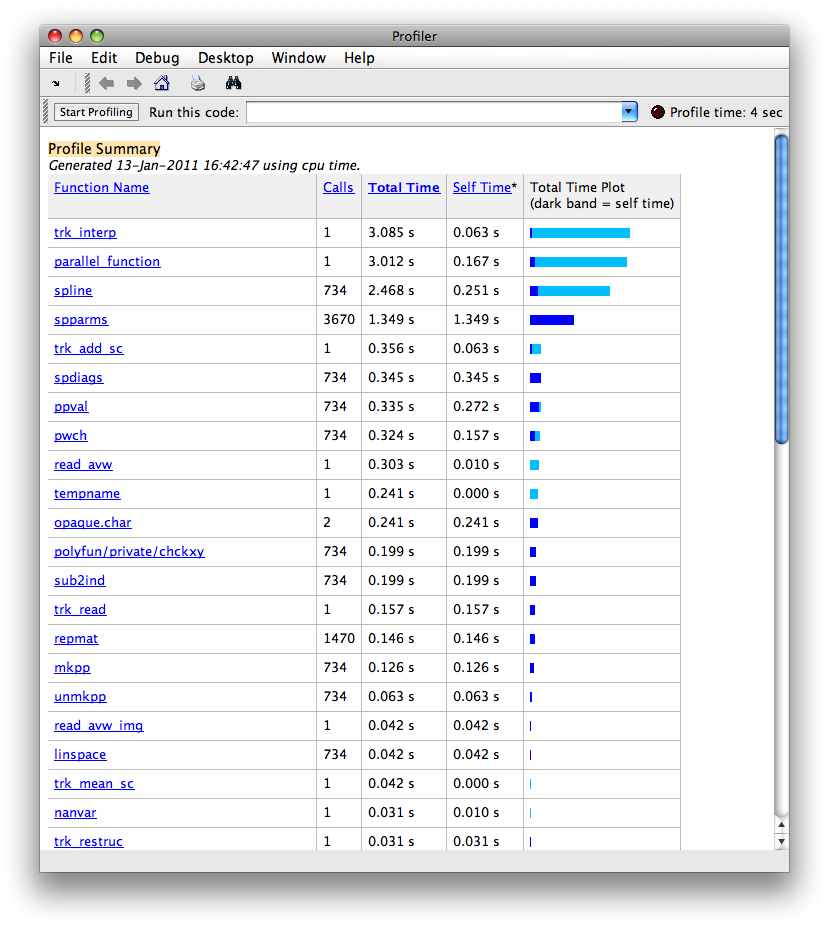
Since most of the time is being spent fitting the splines and resampling the streamlines, you can get a nice boost in speed if you parallelize this step using a parallel for loop (parfor in the Parallel Computing Toolbox).
profile on
tic
trkPath = fullfile(subDir, 'CST_L.trk');
volPath = fullfile(subDir, 'dti_fa.nii.gz');
[header tracks] = trk_read(trkPath);
volume = read_avw(volPath);
tracks_interp = trk_interp(tracks, 100);
tracks_interp = trk_flip(header, tracks_interp, [97 110 4]);
tracks_interp_str = trk_restruc(tracks_interp);
[header_sc tracks_sc] = trk_add_sc(header, tracks_interp_str, volume, 'FA');
[scalar_mean scalar_sd] = trk_mean_sc(header_sc, tracks_sc);
toc
profile viewer
matlabpool closeOutput:
Starting matlabpool using the 'local' configuration ... connected to 7 labs.
Elapsed time is 1.238640 seconds.
Sending a stop signal to all the labs ... stopped.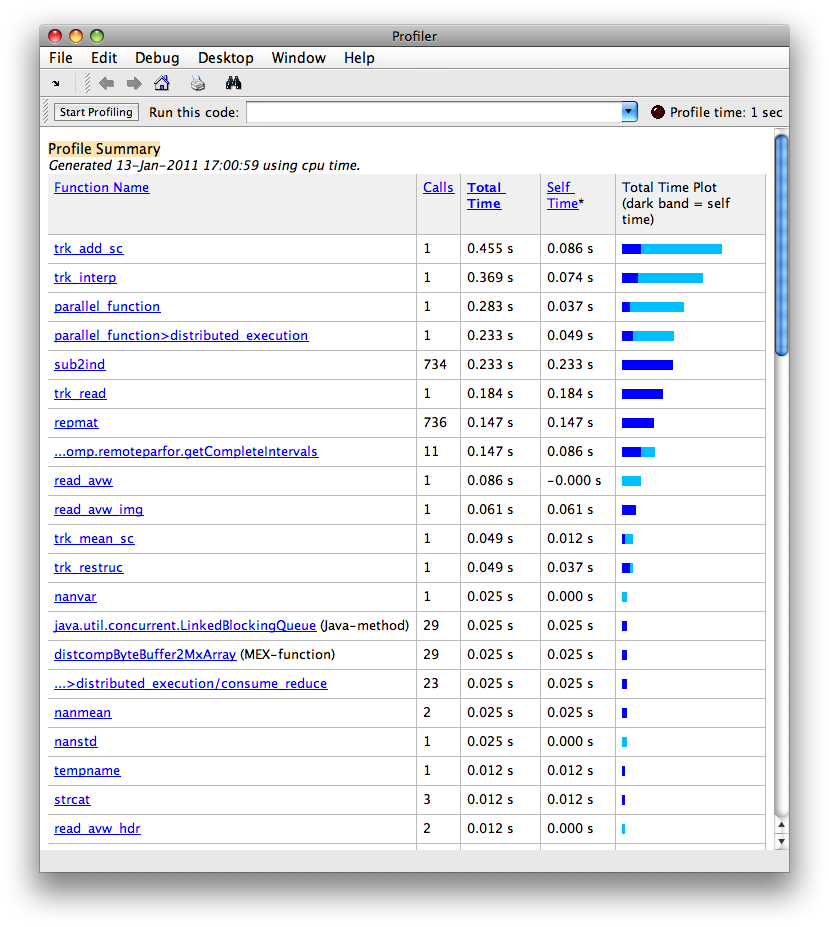
The processing modules that we’ve seen so far can form the core of a loop as we process many subjects and tracts. A bit of pseudo-code looks like this:
loop over subjects
loop over tracts
loop over hemispheres
do along-tract processing steps
end
end
endSee trk_compile_data for an example of a wrapper function that uses this idea.
Back to Wiki Home