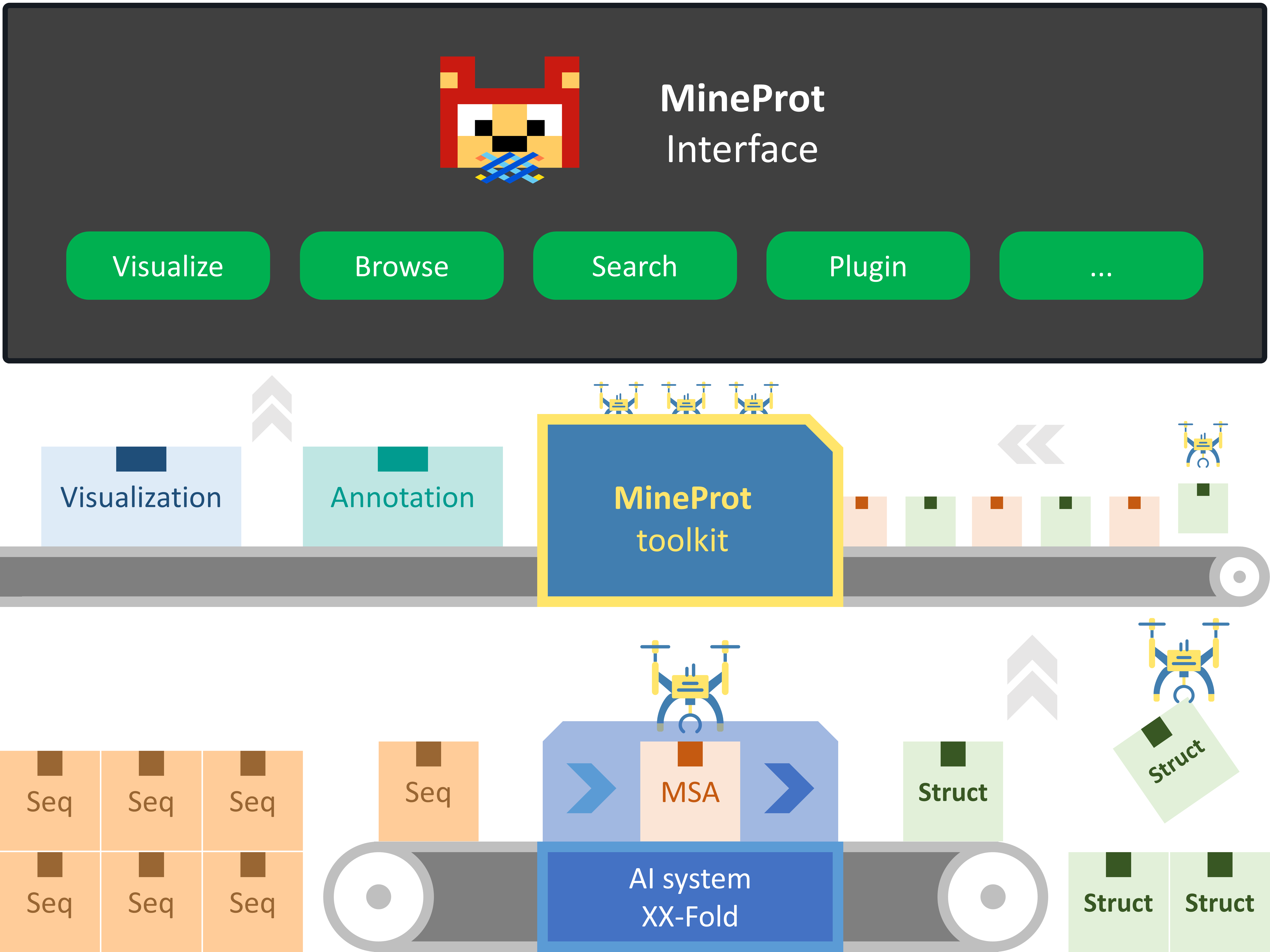
MineProt provides solution for curating high-throughput structuring data from AI systems such as AlphaFold, ColabFold, etc.
By the aid of MineProt toolkit, you can:
- Deploy your own protein server in simple steps
- Take advantage of most information provided by AI systems to curate your data
- Visualize, browse or search your data via user-friendly online interface
- Utilize plugins to extend the functionality of your server
Linux systems with docker and docker-compose are best for MineProt deployment:
git clone https://github.com/huiwenke/MineProt.git
cd MineProt
toolkit/setup.sh -p 80 -d ./dataAfter a few minutes, you can access your MineProt site at http://localhost.
Above all, please get your python3 ready and install dependencies:
cd toolkit/scripts
pip3 install -r requirements.txtThen, use scripts in MineProt toolkit to import your data. For example, if you employ colabfold_search and colabfold_batch with parameters --zip --amber to generate large scale structure predictions, run the command below to import:
colabfold/import.sh /path/to/your/results --repo demo \
--name-mode 1 \
--zip \
--relax \
--python /usr/bin/python3 \
--url http://localhostYou will finally find the protein repository demo at the homepage of your MineProt site, where all proteins have been annotated with UniProt, GO and InterPro, and their structures are visible online.
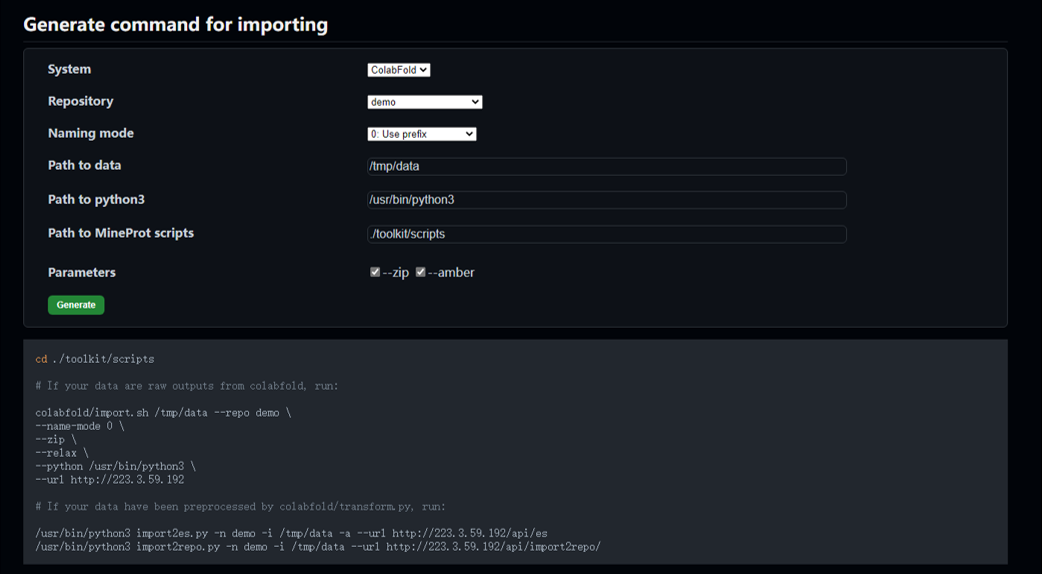
The import page is designed to generate commands for importing, especially useful when you are new to MineProt scripting.
During the import process, PDB structure files are converted into CIF format, which can be visualized by Mol* in the style of AlphaFold DB, while MSAs (A3M files) are used for UniProt annotation and added as keywords in Elasticsearch index. Model scores such as pLDDT and PAE are stored in JSON format.
MineProt interface provides final users with following functions:
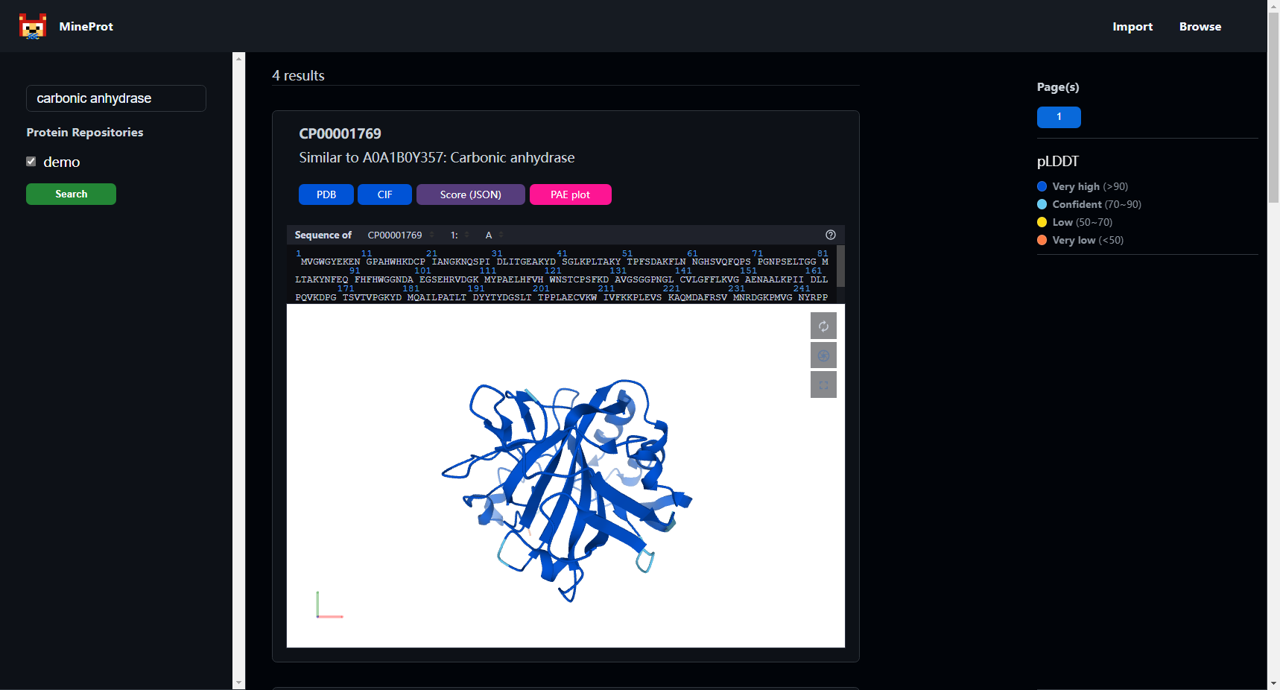
- Search: Type, enter, results.
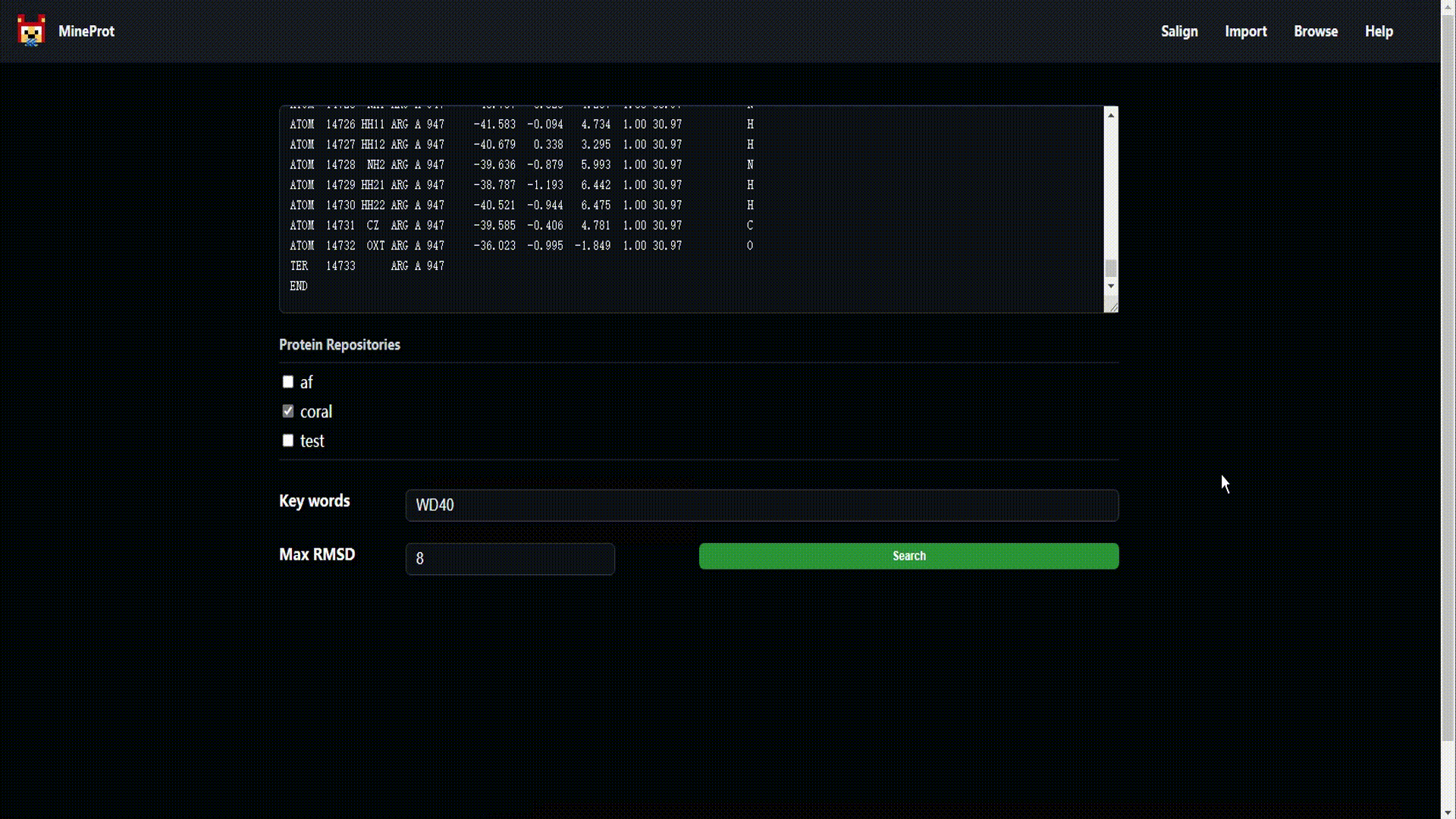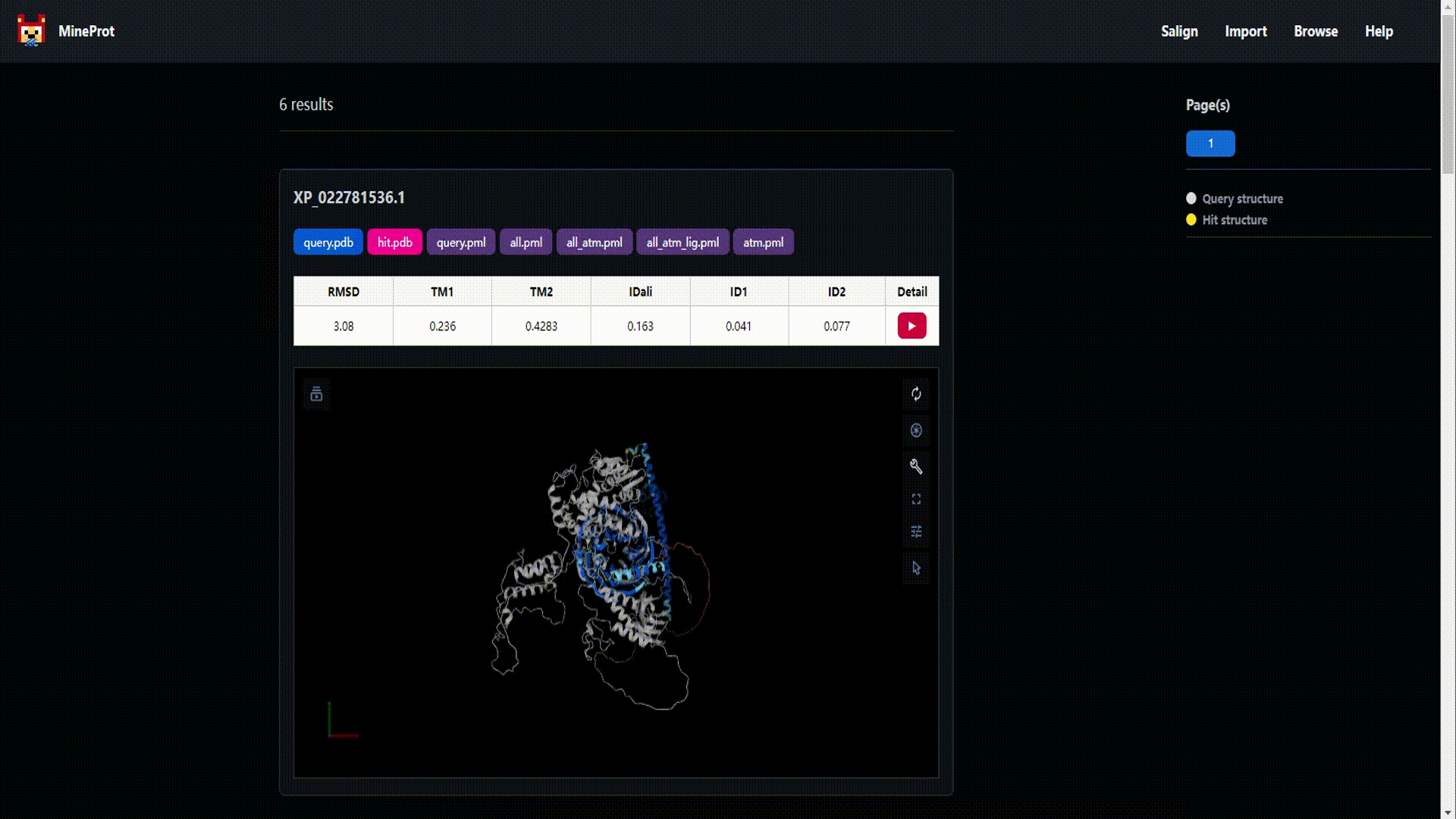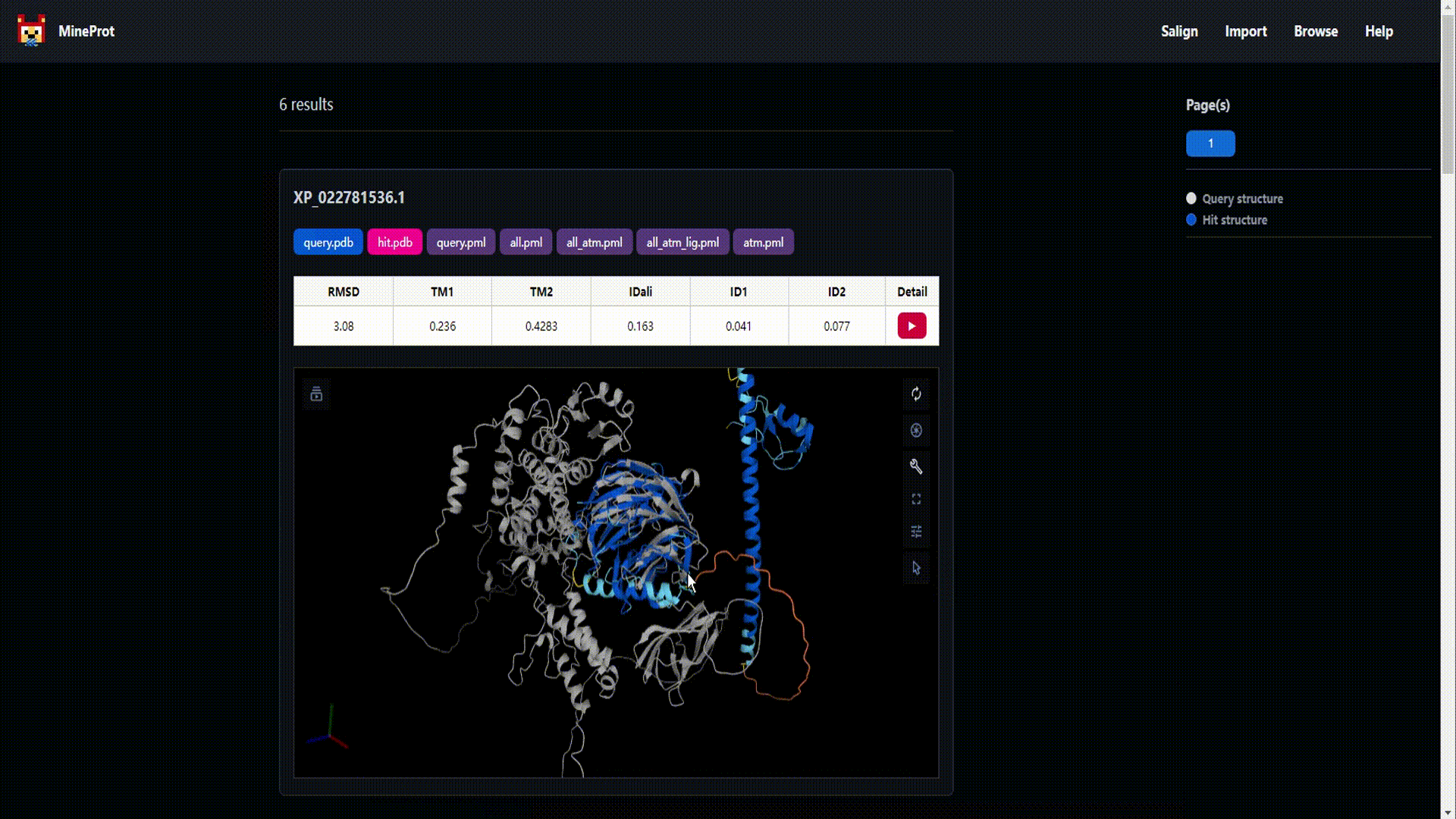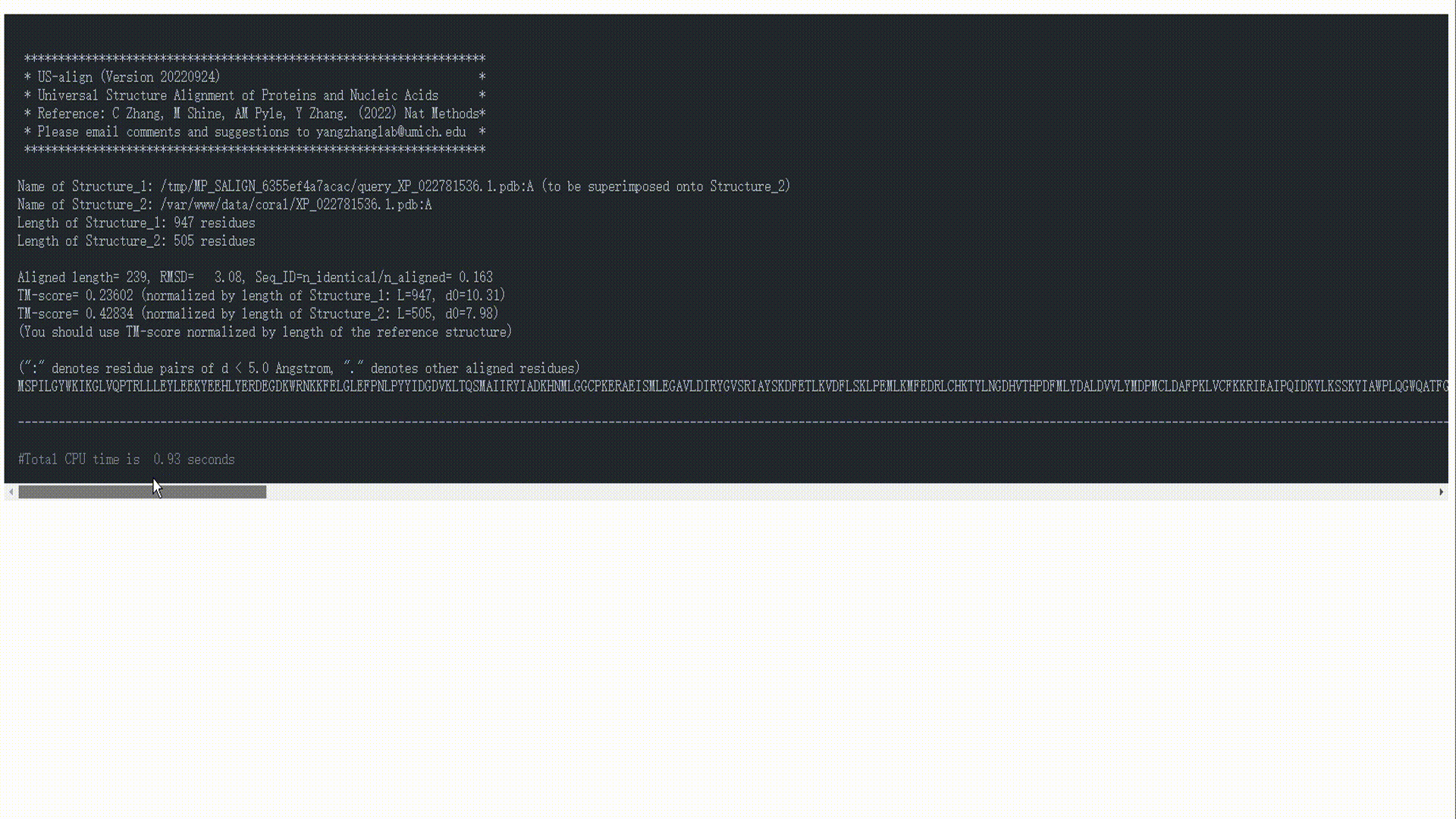
- Visualization: Mol* powered 3D analysis in your browser.
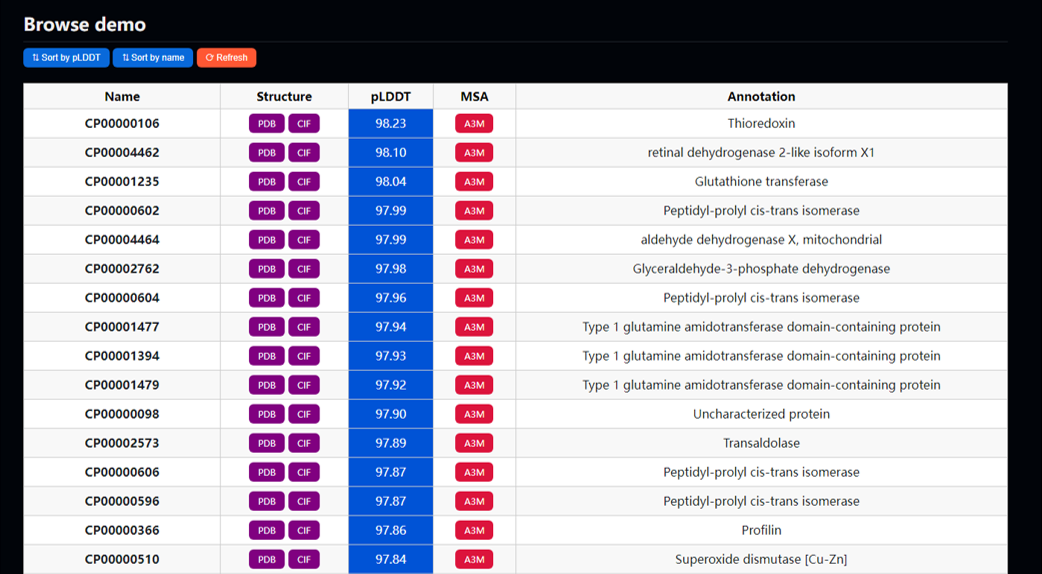
- Browse: Select one repository, and all information will be available.
To speed up response of the browse page, MineProt automatically generates cache for each repository on first access. Clicking the refresh button will regenerate the cache.
- Salign: Structure search engine powered by US-align.
For server administrators, MineProt interface can generate scripts for data importing with simple clicking and copy-pasting in a few steps.
We have tested this project on the following browsers:
| Chrome | Firefox | Edge | Opera | Safari |
|---|---|---|---|---|
| 67.0+ | 52.9+ | 105.0.1343.27 | 90.0.4480.107 | 13.04 |
This project does not support any version of Internet Explorer.
We suggest accessing the web site using devices with resolution higher than 1366x768 (landscape screen) for better user experience.
Usability from mobile devices with portrait screen is not guaranteed.
We recommend Linux platform for non-docker deployment, as MAXIT (converter app between PDB and CIF) is not supported on Windows.
In our developing & testing environment, we employed BioLinux 8.0.7 (PHP 5.5.9, Apache 2.4.7) for non-docker deployment, and Ubuntu 18.04 LTS (Docker 20.10.16, docker-compose 1.29.2) for docker-based deployment.
If you want to deploy this project on your server without Docker, please note that this project and its components depend on the following packages and libraries:
PHP 5.5.9+ with cURL
Elasticsearch 7.12.1
WebServer basic platform (NGINX, Apache, etc.)
MAXIT 11.100
US-align (Version 20220924)
See Deployment manual for more information.
This project's inbox scripts depends on Python 3.6+. Operations will be more convenient with a Bash shell.
Browser plugins require the TamperMonkey plugin.
See Toolkit manual for more information.
@article{10.1093/database/baad059,
title={MineProt: a stand-alone server for structural proteome curation},
author={Zhu, Yunchi and Tong, Chengda and Zhao, Zuohan and Lu, Zuhong},
journal={Database},
volume={2023},
pages={baad059},
year={2023},
publisher={Oxford University Press}
}