-
Notifications
You must be signed in to change notification settings - Fork 10
GenPhyloData Tutorial
JPrIME-GenPhyloData is a suite of tools to generate realistic synthetic (a.k.a artificial or simulated) phylogenetic trees. The models used conform well with the inference tools included in JPrIME, but also support other popular models. Currently, the suite comprises 3 tools:
- HostTreeGen - for generating a bifurcating tree from scratch, typically a species tree.
- GuestTreeGen - for generating a bifurcating tree that evolves w.r.t. a given host tree. This is suitable for simulating gene family evolution.
- BranchRelaxer - for relaxing on the molecular clock assumptions of (1) or (2).
Additionally, as a 4th stage, it should be easy to use an external tool for creating realistic artificial sequences for the host or guest tree leaves (i.e. taxa or genes).
The tools of GenPhyloData are described seperately below.
GenPhyloData is not distributed as a binary, but is included in the JPrIME JAR file, which can be obtained from the download page.
GenPhyloData has been published here. If you use it, please cite:
- Sjöstrand J, Arvestad L, Lagergren J, Sennblad B (2013) GenPhyloData: Realistic simulation of gene family evolution. BMC Bioinformatics. 14:209.
HostTreeGen is a tool to generate bifurcating tree topologies according to a birth-death process. The user inputs a time interval over which the process should take place, along with birth and death rates. Lineages which fail to reach the end of the interval are pruned away. There are additional options regarding, e.g:
- Desired number of leaves.
- Taxon sampling probability.
- Overriding stem edge time (i.e., the edge predating the root split).
- Leaf sizes drawn uniformly (with replacement) from a list of samples. This is convenient to mimic biological data more closely.
- Requiring an immediate split at the start of the time interval.
- Number of attempts to generate a tree that meets the requirements before surrendering.
By default, files are produced for both the unpruned and the pruned tree:
- the tree itself in Newick format.
- an info file detailing the process.
Since both the time interval and the rates are specified, the user can use an arbitrary time unit (e.g. millions of years). The general pattern for the tool is thus:
java -jar jprime-x.y.z.jar HostTreeGen [options] <timespan> <birth rate> <death rate> <out prefix>
To execute the application over time 10, with a birth rate of 0.3 and death rate of 0.1 events per time unit and lineage respectively, and having at least 3 leaves, type the following:
java -jar jprime-x.y.z.jar HostTreeGen -min 3 10 0.3 0.1 myhost
You can always show the list of options by typing:
java -jar jprime-x.y.z.jar HostTreeGen -h
The produced trees are rooted. You may of course use an external tool to remove the root to obtain unrooted trees.
To force the process to start with a split, use option -bi:
java -jar jprime-x.y.z.jar HostTreeGen -bi -min 3 10 0.3 0.1 myhost
This will also force the produced tree to retain this split after pruning, i.e., there will be at least two sampled leaves.
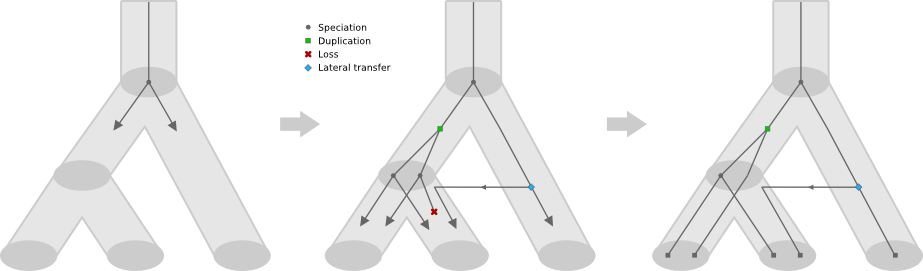
GuestTreeGen is a tool to generate bifurcating "guest" trees that evolve over a known "host" tree according to a birth-death-like process. This is particularly well suited for creating realistic artificial gene families in light of a known taxonomy. The user inputs the host tree over which the process should take place, along with duplication, loss, and lateral transfer rates. The host tree must be rooted, bifurcating, and be dated with ultrametric (clock-like) branch lengths. Guest tree lineages which fail to reach the leaves of the host tree are pruned away. There are a plethora of options, such as:
- Min/max desired number of guest leaves.
- Min/max desired number of guest leaves per host leaf.
- Guest leaf sampling probability.
- Guest leaf sizes drawn uniformly (with replacement) from a list of samples. This is convenient to mimic biological data more closely.
- Number of attempts to generate a tree that meets the requirements before surrendering.
By default, files are produced for both the unpruned and the pruned tree:
- the guest tree itself in Newick format.
- an info file detailing the process.
- a detailed guest-to-host map.
- a guest-to-host map with leaf names only.
Since both the time interval and the rates are specified, the user can use an arbitrary time unit (e.g. millions of years). The general pattern for the tool is thus:
java -jar jprime-x.y.z.jar GuestTreeGen [options] <host tree> <duplication rate> <loss rate> <transfer rate> <out prefix>
where refers to either a string with a Newick tree or a file containing a Newick tree, and refers to the filename prefix of the generated files.
As an example, the following generates a guest tree over a known host tree, with a duplicaton rate of 0.2, loss rate of 0.1, and transfer rate of 0.05 events per time unit and lineage respectively, at least 4 extant guest leaves, and at most 20 extant guest leaves:
java -jar jprime-x.y.z.jar GuestTreeGen -min 4 -max 20 "((A:4.0,B:4.0):6.0,C:10.0):5.0;" 0.2 0.1 0.05 myguest
You can always show the list of options by typing:
java -jar jprime-x.y.z.jar GuestTreeGen -h
The generated guest-to-host leaf map may look like this for a vertebrate host tree (Gx refers to the guest tree leaf names):
G25 vv_tnigroviridis3
G0 mm_tguttata4
G1 mm_oanatinus4
G11 vv_acarolinensis3
G21 mm_oanatinus4
G3 mm_cfamiliaris2
G4 vv_tnigroviridis3
G6 mm_hsapiens5
G7 mm_mdomestica2
G12 mm_ggallus2
G13 vv_tnigroviridis3
G19 mm_mdomestica2
G16 mm_hsapiens5
G17 vv_acarolinensis3
The trees produced in (1) and (2) are ultrametric (clock-like). In order to obtain more realistic branch lengths, this application enables relaxing the molecular clock by rescaling the lengths by heterogeneous rates. A number of popular techniques are supported, including:
- Constant rescaling of the clock-like lengths.
- Uncorrelated models for IID rates drawn from gamma, log-normal and other distributions.
- Rates drawn uniformly (with replacement) from a list of samples. This is convenient to mimic rates from a posterior distribution stemming from a biological MCMC analysis.
- Autocorrelated rates in accordance with Rannala-Yang '07, Thorne-Kishino '98 and some other popular models.
- Host tree guided rates in accordance with Rasmussen-Kellis '11.
- It is possible to constrain the the rates to stay within a desired range, regardless of the model employed.
The input tree must be rooted and bifurcating. Currently, models are employed in a "pure" modeling sense in that rates that concern the edges around the root follow the model without ad hoc constraints. It is otherwise common to, during inference, introduce root-constraints to avoid over-parameterization. However, while the latter may be required for computational reasons, it can hardly be argued as sound behavior during the generative process.
To execute the application, the general pattern is the following:
java -jar jprime-x.y.z.jar BranchRelaxer [options] <tree> <model> <arg1> <arg2> <...>
where <tree> refers to either a string with a Newick tree or a file containing a Newick tree.
You can always show the list of supported rate models and options by typing:
java -jar jprime-x.y.z.jar BranchRelaxer -h
As an example, the following outputs lengths by applying log-normal IID rates, and outputs the relaxed tree to stdout:
java -jar jprime-x.y.z.jar BranchRelaxer "(A:0.5,B:0.5):0.3;" IIDLogNormal 0.0 1.0
If you instead direct output to a file myfile.tree, you will also obtain an additional file detailing the settings, etc. named myfile.tree.info:
java -jar jprime-x.y.z.jar BranchRelaxer -o myfile.tree "(A:0.5,B:0.5):0.3;" IIDGamma 2.0 0.5
It is of course possible to perform multiple rounds of relaxation, should you wish to acquire a combination of models. For instance, you could apply host tree-guided rates first, then relax the lengths further by an IID process.
However, some of the included models (in particular the autocorrelated ones) assume that the input lengths be clock-like, for instance the following, which uses the Thorne-Kishino '98 rate model:
java -jar jprime-x.y.z.jar BranchRelaxer "(A:0.5,(B:0.2,C:0.2):0.3):0.7;" ACTK98 1.0 0.05
The host tree guided rates constitute a particular case, in which one may specify a gamma distribution for every host tree arc as in Rasmussen-Kellis '11. The rate of a guest tree branch is then determined by the distributions of all the host tree edges the branch passes over. The gamma parameters are stated in the host tree like so: [&&PRIME PARAMS=(<k>, <theta>)]. To create such lengths, one would type for example:
java -jar jprime-x.y.z.jar BranchRelaxer g.tree IIDRK11 s.tree gs.sigma 1.2
where 1.2 is a simple scale factor applied to all lengths after relaxation. The host tree s.tree might look like:
((A:0.5[&&PRIME PARAMS=(10,0.1)],B:0.5[&&PRIME PARAMS=(10,0.1)]):0.4[&&PRIME PARAMS=(5,0.2)],C:0.9[&&PRIME PARAMS=(5,0.2)]):0.3[&&PRIME PARAMS=(10,0.1)];
and the guest tree g.tree might look like:
((a1:0.2,a2:0.2):0.8,(b1:0.9,c1:0.9):0.1):0.2;
and the file gs.sigma linking the trees together might look like:
a1 A
a2 A
b1 B
c1 C
Note however, that the host tree and guest tree must be ultrametric and temporally compatible, and that the model is currently only applicable on guest trees which lack lateral transfer events.
If you wish to produce synthetic sequence data for your trees, you may use, e.g., Seq-Gen (Rambaut, 2002), which supports a wide range of popular models of molecular sequence evolution for nucleotide, amino acid or codon data. Such programs typically support modelling also indels, gaps, and site rate heterogeneity.
A more recent application to model whole genomes at a time is ALF (Dalquen et al., 2012).
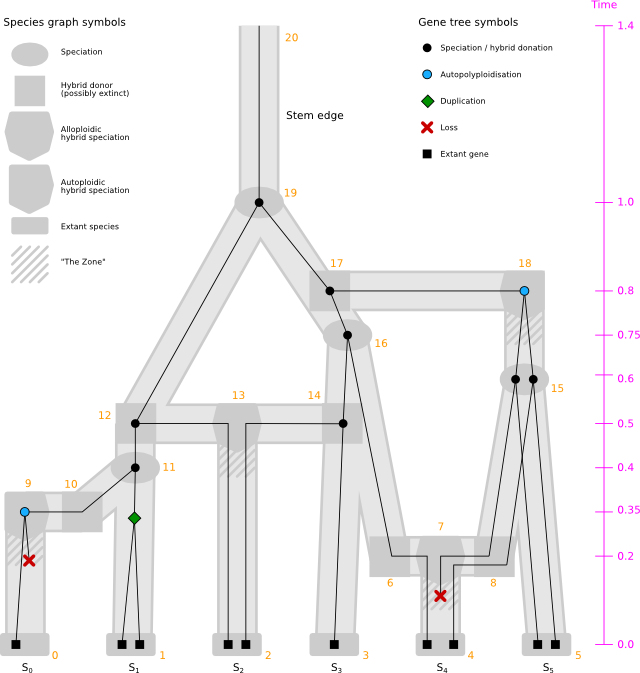
GuestTreeGen has been extended with the ability to generate bifurcating guest trees with duplication and loss events over a host graph with host vertices representing alloploidic and autoploidic hybrid speciations. The graph is to be provided on the GML format, such as the one shown above: hybrid_graph_w_extinctions.gml. The process works as usual, albeit with the following remarks:
- The transfer rate is ignored (i.e., set to 0).
- For a guest lineage reaching an autoploidic host vertex, an obligate duplication occurs.
- The model supports extinct hybrid donor species as well.
- It is possible to specify a different duplication and loss rate following a hybrid speciation. This is handled by defining factors with which the rates are rescaled for a certain amount of time. This allows for temporarily increasing the loss rate, for instance.
- The unpruned guest tree may have vertices with outdegree 1 to indicate paths over hybrid host vertices. The pruned guest tree is bifurcating, however.
You specify the hybrid graph option in the following manner:
java -jar jprime-X.Y.Z.jar GuestTreeGen -hybrid 0.05 1 2 hybrid_graph_w_extinctions.gml 0.1 0.005 0 myhybridguest
to generate a tree where the post-hybridisation "zone" is set to last for 0.05 time units, with a duplication rate temporarily rescaled by 1 and the loss rate temporarily rescaled by 2.
Q1) There seems to be a small discrepancy between the times of my host tree and the times of the generated guest tree vertices that are placed onto host tree vertices. Why?
A1) If the difference is really miniscule, surely this is due to internal numerical precision effects. However, should it be larger (say in the order of 1e-4), this is most likely due to that there are internal host tree vertices with equal times. The time-colliding vertices have then been moved slightly to accomodate for non-ambiguous transfer events. This will happen even if you set the transfer rate to 0.