3DCT_for_PKU_LDG is modified from 3DeeCellTracker for Luo_lab fly transcriptional signal extraction.
Modifications include:
- CellTracker/unet3d.py line 36: change "inputs = Input((160,160,16,1))" to "inputs = Input((80,80,8,1))". Luo-lab pic size is 460*157, smaller than 160
- CellTracker/tracker.py line 1258: change "self.segresult.r_corrdinates_segment, 20" to "self.segresult.r_corrdinates_segment, 5" Luo-lab cell number per volume is less than 20. Note that the ffn model should also be updated accordingly
- CellTracker/analyses.py line 15-88: define a get_signals function. Used for singal extraction
- CellTracker/tracker.py line 752: add "self.raw_data = _make_folder(os.path.join(folder_path, "raw_data/"))" to create a folder for storing raw data We didnt pre-align the raw data, so we use raw_data folder to store these images, the reason is to keep the orginal folder structure(using the data folder for aligned data)
- CellTracker/tracker.py line 752: add "self.cell_trace = _make_folder(os.path.join(folder_path, "cell_trace/"))" to create a folder for storing final cell traces The folder is used for storing final results
Examples/signal_extraction.ipynb: a jupyter notebook file for signal extraction.- Examples/single_mode_worm1-clear.ipynb: integrate the signal extraction operation.
- 3DCT.yml line 18: comment out to create a conda env which can be installed with modified site package (CellTracker).
Installation
$ git clone https://github.com/vislabwwy/3DCT_for_PKU_LDG.git
$ cd 3DCT_for_PKU_LDG
$ conda env create -f 3DCT.yml -n 3DCT_PKU_LDG
$ conda activate 3DCT_PKU_LDG
$ python setup.py install3DeeCellTracker is a deep-learning based pipeline for tracking cells in 3D time lapse images of deforming/moving organs (eLife, 2021).
3DeeCellTracker v0.4.5 was released with following issues fixed (2022.06.03)
- Solved an activity load bug.
- Create a conda environment for a PC with GPU including prerequisite packages using the 3DCT.yml file:
$ conda env create -f 3DCT.yml- (NOT RECOMMEND) Users can create a conda environment for a PC with only CPU, but it will be slow and may fail.
$ conda env create -f 3DCT-CPU.yml- Install the 3DeeCellTracker package solely by pip
$ pip install 3DeeCellTracker- Update the 3DeeCellTracker package to the latest version
$ pip install --upgrade 3DeeCellTrackerFor detailed instructions, see here.
To learn how to track cells use 3DeeCellTracker, see following notebooks for examples:
-
Track cells in deforming organs:
-
Track cells in freely moving animals:
-
Train a new 3D U-Net for segmenting cells in new optical conditions:
The data and model files for demonstrating above notebooks can be downloaded here.
Note: Codes above were based on the latest version. For old programs used in eLife 2021, please check the "Deprecated_programs" folder.
Multiple users have reported that when running the tracker.match() function, a ValueError of shape mismatch occurs. And they were discovered to be the result of an incorrect setting of siz_xyz, which should be (height, width, depth) of the 3D image. If you encounter the same error, please double-check that it is correctly set.
We have made tutorials explaining how to use our software. See links below (videos in Youtube):
Tutorial 1: Install 3DeeCellTracker and train the 3D U-Net
Tutorial 2: Tracking cells by 3DeeCellTracker
Tutorial 3: Annotate cells for training 3D U-Net
Tutorial 4: Manually correct the cell segmentation
We have wrote a tutorial explaining how to install and use 3DeeCellTracker. See Bio-protocol, 2022
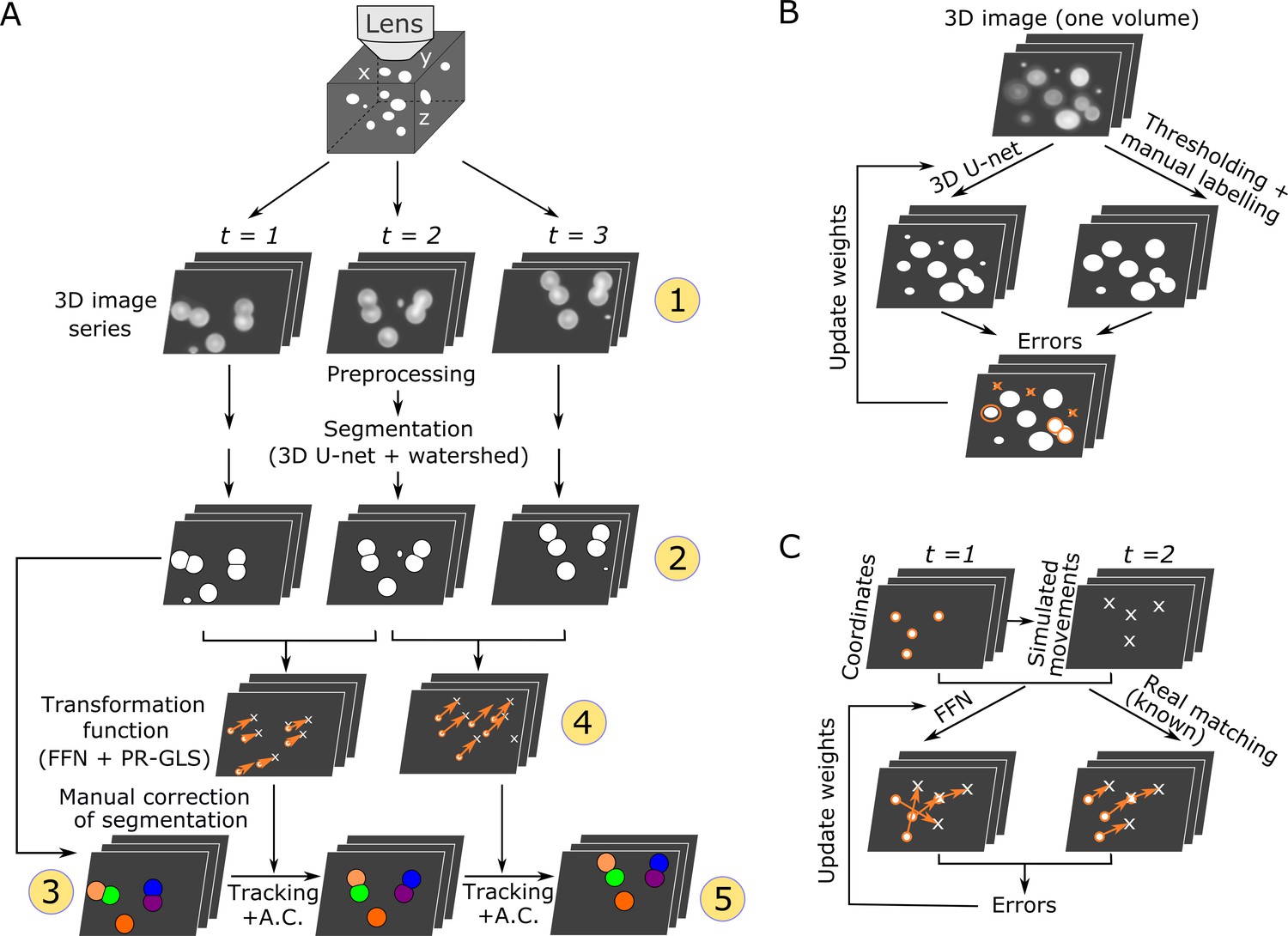
We designed this pipeline for segmenting and tracking cells in 3D + T images in deforming organs. The methods have been explained in Wen et al. bioRxiv 2018 and in Wen et al. eLife, 2021.
Overall procedures of our method (Wen et al. eLife, 2021–Figure 1)
Examples of tracking results (Wen et al. eLife, 2021–Videos)
| Neurons in a ‘straightened’ freely moving worm |
Cardiac cells in a zebrafish larva | Cells in a 3D tumor spheriod |
|---|---|---|
 |
 |
 |
If you used this package in your research and is interested in citing it here's how you do it:
@article{
author = {Wen, Chentao and Miura, Takuya and Voleti, Venkatakaushik and Yamaguchi, Kazushi and Tsutsumi, Motosuke and Yamamoto, Kei and Otomo, Kohei and Fujie, Yukako and Teramoto, Takayuki and Ishihara, Takeshi and Aoki, Kazuhiro and Nemoto, Tomomi and Hillman, Elizabeth MC and Kimura, Koutarou D},
doi = {10.7554/eLife.59187},
journal = {eLife},
month = {mar},
title = {{3DeeCellTracker, a deep learning-based pipeline for segmenting and tracking cells in 3D time lapse images}},
volume = {10},
year = {2021}
}
We wish to thank JetBrains for supporting this project with free open source Pycharm license.