-
Notifications
You must be signed in to change notification settings - Fork 0
B. Anat and Functional Pre processing
To start working on this project, make sure you have downloaded the anatomical and functional resting-state scans of the MPI_Lemon dataset to your computer. You will also need to have the behavioral data.
You will need to set the location of the data in basics.py. In particular make sure to provide the correct values to variables ORIG_FMRI_DIR and ORIG_BEHAV_DIR.
The first notebook in this project is S00_OrganizeData. This notebook will inspect the downloaded dataset and generate three outputs:
-
NC_withSNYCQ_subjects.txt: List of subjects with resting data that has a completed SNYCQ dataset and save it to -
SNYCQ_Preproc.pkl: Dataframe with all the responses to the SNYCQ. In this dataframe, rows corresponds to different runs and columns correspond to each of the SNYCQ questions.NOTE: To ensure this repo does not include any subject data, this framework gets written outside the directory structure of the repo. Make sure you created the target folder in advance.
-
NC_anat_info.csv: Dataframe with information about when anatomical data was acquired for each subject (e.g., ses-01 or ses-02). This dataframe will be used in STEP02 to generate the correct calls to the structural pipelines.
Runs all cells in this notebook, check the printed outputs and ensure the three primary output files are saved to disk.
- Create the output folder for Freesurefer
mkdir /data/SFIMJGC_Introspec/pdn
mkdir /data/SFIMJGC_Introspec/pdn/Freesurfer
-
Run all the cells in notebook
S01_NC_run_structural.CreateSwarmto generate a swarm fileS01_NC_run_structural.SWARM.shin the swarm folder. This swarm file will contain one call per subject to the structural pre-processing pipeline. -
Submit the swarm file to the cluster
swarm -f /data/SFIMJGC_Introspec/2023_fc_introspection/SwarmFiles.<your_user_name>/S01_NC_run_structural.SWARM.sh -g 32 -t 32 --time 32:00:00 \
--logdir /data/SFIMJGC_Introspec/2023_fc_introspection/Logs.<your_user_name>/S01_NC_run_structural.logs
NOTE: As part of this pipeline there is a call to Freesurfer recon-all, and to ANTs registration. This means that this step can take approx. 9 hours to run on each subject. Sit, relax, and pick your favorite book or paper to read.
Description of the structural pre-processing as in Mendes et al. 2009:
"Structural data. The background of the uniform T1-weighted image was removed using CBS Tools, and the masked image was used for cortical surface reconstruction using FreeSurfer’s full version of recon-all. A brain mask was created based on the FreeSurfer segmentation results. Diffeomorphic nonlinear registration as implemented in ANTs SyN algorithm was used to compute a spatial transformation between the individual’s T1-weighted image and the MNI152 1mm standard space. To remove identifying information from the structural MRI scans, a mask for defacing was created from the MP2RAGE images using CBS Tools. This mask was subsequently applied to all anatomical scans."
Once all anatomical pre-processing jobs are completed, you should check for errors. In our case, we found that anatomical preprocessing failed for 25 subjects due to reasons such as: hypersensitivity in white matter, errors in skull stripping, etc. A list of the subjects with errors can be found in resources/preprocessing_notes/NC_struct_fail_list.csv
-
Run all the cells in notebook
S02_NC_run_func_preproc.CreateSwarmto generate a swarm fileS02_NC_run_func_preproc.SWARM.sh -
Submit the swarm file to the cluster
swarm -f /data/SFIMJGC_Introspec/2023_fc_introspection/SwarmFiles.<your_user_name>/S02_NC_run_func_preproc.SWARM.sh -g 32 -t 32 --time 32:00:00 \
--logdir /data/SFIMJGC_Introspec/2023_fc_introspection/Logs.<your_user_name>/S02_NC_run_func_preproc.logs
NOTE: This step uses afni, fsl and freesurfer. It takes approx. 5 hours to run.
Description of the functional pre-processing pipeline as in Mendes et al. 2009:
"Functional data. The first five volumes of each resting-state run were excluded. Transformation parameters for motion correction were obtained by rigid-body realignment to the first volume of the shortened time series using FSL MCFLIRT79. The fieldmap images were preprocessed using the fsl_prepare_fieldmap script. A temporal mean image of the realigned time series was rigidly registered to the fieldmap magnitude image using FSL FLIRT80 and unwarped using FSL FUGUE81 to estimate transformation parameters for distortion correction. The unwarped temporal mean was rigidly coregistered to the subject’s structural scan using FreeSurfer’s boundary-based registration algorithm82, yielding transformation parameters for coregistration. The spatial transformations from motion correction, distortion correction, and coregistration were then combined and applied to each volume of the original time series in a single interpolation step. The time series were masked using the brain mask created from the structural image (see above). The six motion parameters and their first derivatives were included as nuisance regressors in a general linear model (GLM), along with regressors representing outliers as identified by Nipype’s rapidart algorithm (https://nipype.readthedocs.io/en/latest/interfaces/ generated/nipype.algorithms.rapidart.html), as well as linear and quadratic trends. To remove physiological noise from the residual time series, we followed the aCompCor approach as described by Behzadi and colleagues83. Masks of the white matter and cerebrospinal fluid were created by applying FSL FAST84 to the T1-weighted image, thresholding the resulting probability images at 99%, eroding by one voxel and combining them to a single mask. Of the signal of all voxels included in this mask, the first six principal components were included as additional regressors in a second GLM, run on the residual time series from the first GLM. The denoised time series were temporally filtered to a frequency range between 0.01 and 0.1 Hz using FSL, mean centered and variance normalized using Nitime85. The fully preprocessed time series of all for runs were temporally concatenated. To facilitate analysis in standard space, the previously derived transformation was used to project the full-length time series into MNI152 2 mm space."
NOTE: Although we run the whole Mendes et al. functional pre-processing pipeline, we will gather its outputs following the last spatial transformation step (in bold above) and continue pre-processing with our own AFNI code. The reasons for this approach include: 1) computational efficiency, 2) performance of all nuisance regression in a single GLM step, 3) normalization of time-series to signal percent change.
NOTE: Once all these jobs were concluded we checked the data and we observed that a few scans had not completed. A list of those scans and the reasons for incorrect completion is listed in
resources/preprocessing_notes/NC_func_fail_list.csv
In addition to checking the correct completion of the structural and functional pipelines, we also discarded scans based on excessive motion. In particular, for every volume with a relative motion above 0.2mm we marked as such volumes, the one before and the two subsequent as volumes to be censored in subsequent analyses.
Any scan that had 30% or more of the volumes marked as censored volumes was discarded from further analysis.
The notebook that performs this motion QA is S03_QA_ExcessiveMotion.
A list of scans removed from further analyses due to having excessive motion can be found in file resources/preprocessing_notes/NC_func_too_much_motion_list.csv
| Distribution of censored volumes per scan |
|---|
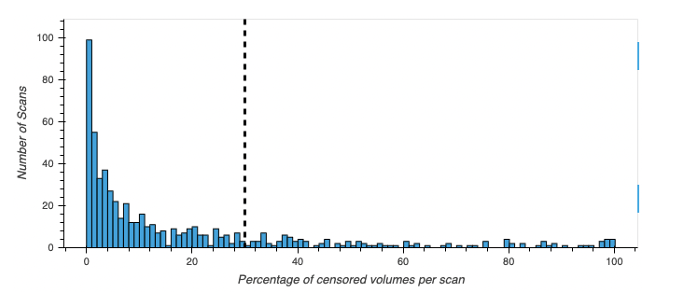 |
Notebook S04_Transform to MNI.CreateSwarm gathers the spatial transformation matrix to go from subject's original space into MNI space and applies them to both the motion correction version of each resting-state scan and also to its temporal mean. The notebook proceeds in three separate steps:
-
S04_TransformToMNI.pass01.sh: Transform the motion corrected (and its mean) output by nipype to MNI space using ANTs (same command used in the original nypipe pipeline). The size of files in MNI is 2.1GB. Because of that, we perform two additional steps that help reduce file size. -
S04_TransformToMNI.pass02.sh: Compute a common grid in MNI space that will include everybody's skull stripped data, but no boundary of zeros around it. As part of this step there are two manual interventions. First, we manually correct the group average intra-cranial mask to remove some extra 1's in right frontal cortex left by AFNI's3dAutomask. Second, we manually generated a rough CSF mask that we use later to remove non-ventricular CSF voxels when generating a mark for obtaining CompCorr regressors. -
S04_TransformToMNI.pass03.sh: Force everybody's resting-state scans in MNI space into the smaller grid (with no surrounding empty voxels). This reduce file size by half to a final size of approx. 1.3 GB.
NOTE: This step required manual intervention at two points. First, to correct the average whole-brain mask due to an extrusion in the right frontal region that is no brain. The corrected version is available in this repo as
all_mean.mask.boxed.nii.gz. Second, we also manually generated a mask that shows the areas to likely contain the ventricles. The mask is in fileall_mean.mask.CSF_region.boxed.nii.gz. This file will be generated later when creating the mask to extract CompCorr regressors.
| Masks that required manual intervention |
|---|
 |
Notebook S05_SegmentT1 will run AFNI's command 3dSeg on the MNI version of the T1 scan of each subject. This will generate probabilistic tissue maps that will be used later (STEP 7) to generate scan specific masks for GM, WM and ventricular compartments.
The notebook creates a swarm file with one entry per subject. The exact call to 3dSeg can be found on bash script S05_SegmentT1.sh
The last pre-processing steps prior to extracting representative timeseries per ROI are temporal scaling and nuissance regression. The notebook in charge of generating the swarm jobs to perform those steps is S06_NuissanceRegression.
For scaling, we transform voxel-wise timeseries into signal percent change units. This is done here.
Regarding, nuissance regression, in this work we used the following regressors:
- Bandpass Regressors [0.01 - 0.15 Hz]: those are generated here.
- Motion and 1st Derivative: those are generated here.
- Slow Trends: we pass polort = 3 to AFNI's 3dDeconvolve call.
- Physiological Noise: for this we rely on the CompCorr method. We first generate tissue specific masks and the extract the first 5 PCA components within the tissue mask. It is this 5 components that represent the physiological noise regressors.
Nuissance regression is accomplished using AFNI program 3dTproject as shown here.
| Example of mask and traces associated with the CompCorr/Physio regression |
|---|
 |