The goal of neuromorphr is to provide R client utilities for interacting with the API for neuromorpho.org. The site neuromorpho.org is a well-known centrally curated inventory of digitally reconstructed neurons associated with peer-reviewed publications. It is continuously updated as new morphological reconstructions are collected, published and shared. It contains contributions from hundreds of laboratories worldwide (see many here). To date, neuromorpho.org is the largest collection of publicly accessible 3D neuronal reconstructions (>100,000) and associated metadata which can be used for detailed single cell simulations. This R package was built to work with version 7.7 of neuromorpho.org.
If python is more your poison, there are a few python clients available, for example from BonsaiNet. Using this R package in concert with the nat ecosystem developed primarily by Gregory Jefferis is highly recommended. The curators of neuromorpho.org can be contacted at [email protected].
In the following, we detail some of neuromorphr's functionality, and copy and paste some text from neuromorpho.org in order to fill you in on its database and outlook.
Firstly, you will need R, R Studio and X Quartz as well as nat and its dependencies. For detailed installation instructions for all this, see here. It should not take too long at all. Then:
# install
if (!require("remotes")) install.packages("remotes")
remotes::install_github("natverse/neuromorphr")
# use
library(neuromorphr)Done!
Now we can have a look at what is available, here are some of the key functions. Their help details examples of their use. You can summon the help in RStudio using ? followed by the function name.
# Sometimes neuromorphr.org can be slow, or down from capacity issues or have a faulty API. Can I check this?
neuromorpho_is_api_healthy()
# And how can I read neurons from neuromorpho?
?neuromorphr_read_neurons()
# But first I need neuron names and/or neuron IDs, how do I get those?
?neuromorpho_search()
# I see, so what metadata do I get with my neurons?
?neuromorpho_neurons_info()
?neuromorpho_neurons_meta()
# Interesting, I see a pubmed ID (pmid) is also given. Can I find the scientific artices that describe these neurons?
?neuromorpho_articles_from_neurons()
?neuromorpho_literature_search()
?neuromorpho_get_article()
# I heard something about persistence vectors, they're used to describe describe meaningful morphological features? Can I get those?
?neuromorpho_persistence_vectorsEach neuron in neuromorpho.org is represented by a name, general information (metadata) and a standardised SWC file for the digital morphological reconstruction. Most neurons also have some basic measurements calculated, including for example cable length, in micrometers, and volume, in micrometers cubed. The functions above can get you this information.
Here is a quick example for reading some neocortical rat, elephant and whale neurons; plotting them and comparing them:
# Let's get neocortical neurons from both the African elephant, the humpback whale and the rat
neocortex.df = neuromorpho_search(search_terms= c("species:elephant,humpback whale,rat", "brain_region:neocortex"))
## Pull measurements, in a data frame
measurements = neuromorpho_morphometry(neocortex.df$neuron_name, batch.size = 2, data_frame = TRUE)
## Assign species column
measurements$species = neocortex.df[rownames(measurements),"species"]
## Boxplot, neuron length
boxplot(as.numeric(length)~species, data=measurements, notch=FALSE,
col=(c("deepskyblue1","firebrick1", "darkolivegreen1")),
main="neocortical neuron lengths", xlab="species")
## Boxplot, neuron volume
boxplot(as.numeric(volume)~species, data=measurements, notch=FALSE,
col=(c("deepskyblue1","firebrick1", "darkolivegreen1")),
main="neocortical neuron volumes", xlab="species")
## Read neurons
pyramidal.cells = neuromorpho_read_neurons(neuron_name = neocortex.df$neuron_name, batch.size = 2, nat = TRUE, progress = TRUE)
## Plot 3D
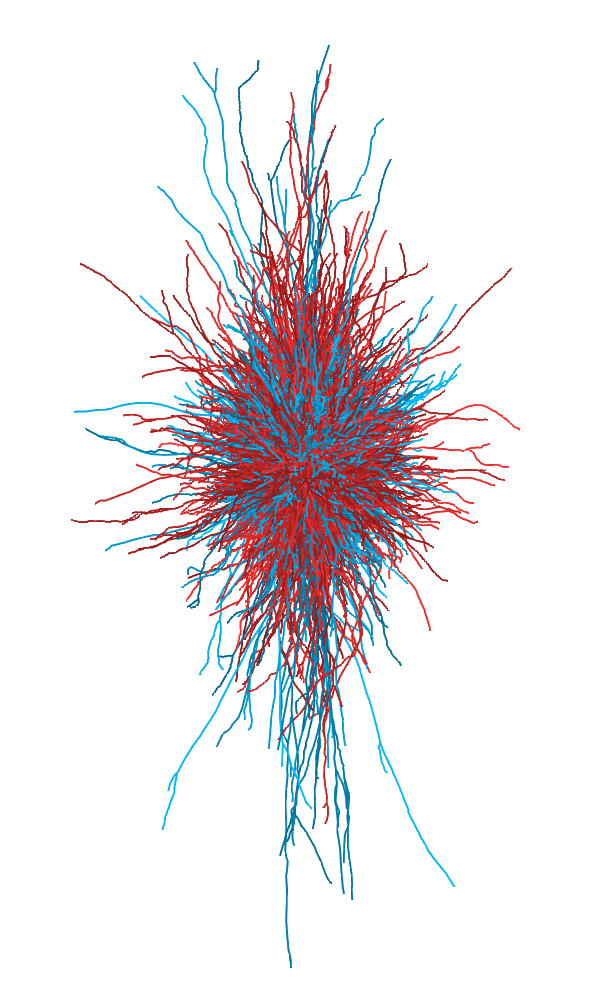
plot3d(pyramidal.cells, col = as.factor(species))Here's a subset. In red, elephant, and in blue, whale, cortical pyramidal neurons:
As well as using this package to interact with neuromorpho.org curated data, you can consider submitted your own neurons if you have been involved in lab work that has acquired 3D reconstructions. The submission process is very straightforward:
- E-mail the reconstruction files (zipped, if possible) to: [email protected]
- Fill in as much information possible in the Metadata Form and include it in your e-mail
Feed the beast.
The neuromorpho.org has a terms of use, which provides guidance on how best to credit data from these repositories. Most neurons have an associated publication that you can fidn using this package or directly on tthe repositroy websites.
This package was created by Alexander Bates, while in the group of Dr. Gregory Jefferis. You can cite this package as:
citation(package = "neuromorphr")Bates AS (2019). neuromorphr: R client utilities for interacting with neuron morphology repositories. R package version 0.1.0. https://github.com/jefferislab/neuromorphr
neuromorpho.org was started and is maintained by the Computational Neuroanatomy Group at the Krasnow Institute for Advanced Study, George Mason University, under the direction of Prof. Giorgio Ascoli, PhD. This project is part of a consortium for the creation of a "Neuroscience Information Framework," endorsed by the Society for Neuroscience, funded by the National Institutes of Health, led by Cornell University (Dr. Daniel Gardner), and including numerous academic institutions such as Yale University (Dr. Gordon Shepherd), Stanford University (Dr. Paul Sternberg), and University of California, San Diego (Dr. Maryann Martone). The neuromorpho.org mission statement can be found here.
Ascoli GA (2006) Mobilizing the base of neuroscience data: the case of neuronal morphologies. Nature Rev. Neurosci., 7:318-324
Ascoli GA, Donohue DE, Halavi M. (2007) NeuroMorpho.Org: a central resource for neuronal morphologies. J Neurosci., 27(35):9247-51