A simple script for creating and maintaining UCSC genome browser track/assembly hubs using google spreadsheets
The UCSC Genome Browser is a web-based genome browser widely used for viewing and sharing various types of genomic data mapped to reference genomes. Track hubs allow users to create and maintain a stable database of custom tracks for viewing in the browser. Additionally, an assembly hub is a type of track hub that allows users to explore other reference genomes not hosted at UCSC. Creating and maintaining multiple hubs with a large number of tracks is difficult and time-consuming because the markup-type language used to define hubs, while allowing for complex track relationships to be defined, becomes difficult to maintain with a large number of tracks.
Track/assembly hubs are entirely defined by a set of files stored by the user on a web server that the UCSC browser can access via http. gtracks is a bash script that allows users to define and maintain multiple track/assembly hubs in a set of google spreadhsheets, which are much easier to maintain by humans for a large number of tracks and hubs. From your web server hosting the data, gtracks downloads these google spreadsheets and processes them into the set of files required to define a track/assembly hub, including creating the entire directory structure for each hub if it isn't already present. Apart from the google spreadsheets, the only other information that needs to be maintained by the user are the files corresponding to the data tracks to be loaded. Below shows an example set of track/assembly hubs. The only files that need to be maintained by the user, which are starred below, are the data files for each track, and in the case of assembly hubs, the 2bit file containing the reference genome sequence.
hubs/ - directory containing a set of track/assembly hubs to be managed by gtracks
gtracks.sh - the gtracks shell script
trackHub1/ - directory containing files for trackHub1
hub.txt - a short description of hub properties
genomes.txt - list of genome assemblies included in the hub data
hg19/ - directory for the hg19 human assembly
trackDb.txt - display properties for tracks in this directory
bbi/ - directory of data files containing the data to be displayed for hg19
*dnaseLiver.bigWig - wiggle plot of DNase in liver
*dnaseLiver.bigBed - regions of active DNase
hg18/ - directory of data for the hg18 human assembly
trackDb.txt - display properties for tracks in this directory
bbi/ - directory of data files containing the data to be displayed for hg18
*dnaseLiver.bigWig - wiggle plot of DNase data in liver
*dnaseLiver.bigBed - regions of active DNase
assemblyHub1/ - directory containing files for assemblyHub1
hub.txt - a short description of hub properties
genomes.txt - list of genome assemblies included in the hub data
newOrg1/ - directory for the newOrg1 assembly
trackDb.txt – definitions for tracks on this genome assembly
*genome.2bit – ‘2bit’ file constructed from your fasta sequence
bbi/ - directory of data files containing the data to be displayed for hg19
*dnaseLiver.bigWig - wiggle plot of DNase in liver
*dnaseLiver.bigBed - regions of active DNase
newOrg2/ - directory for the newOrg2 assembly
trackDb.txt – definitions for tracks on this genome assembly
*genome.2bit – ‘2bit’ file constructed from your fasta sequence
bbi/ - directory of data files containing the data to be displayed for hg19
*dnaseLiver.bigWig - wiggle plot of DNase in liver
*dnaseLiver.bigBed - regions of active DNase
If a user does not have access to such a server, one can easily and cheaply generate one on a cloud service provider such as Digital Ocean.
First download the gtracks script, make it executable, and place it in the directory on your webserver you want to store your track/assembly hubs.
Copy google folder containing spreadsheet templates to your google drive. Make sure the entire folder is shared via link sharing. The folder contains the following sheets:
_hubDb
exampleHub_hub
exampleHub_genomes
exampleHub_trackDb_assembly
Spreadsheets are named using the "name" field specified in the _hubDb spreadsheet, in this case "exampleHub". The "assembly" is specified in the genomes sheet (name_genomes) via the "genome" field. This assembly can either be a UCSC genome assembly id (thus making a track hub) or an id for a provided reference assembly (thus making an assembly hub) in which case the twoBit field must also be specified in the genomes sheet and a 2bit reference genome must be placed on the webserver in the hub/assembly/ directory. Only one _hubDb sheet is required for as many hubs as desired. Only one name_hub and name_genomes sheet is required for each hub. The name_genomes file can contain multiple rows specifiying any combination of track and/or assembly hubs. A name_trackDb_assembly sheet is required for each genome specified in the name_genomes sheet.
Now set the id keys in each spreadsheet according the the following schema:
_hubDb -> name_hub -> name_genomes -> name_trackDb_assembly
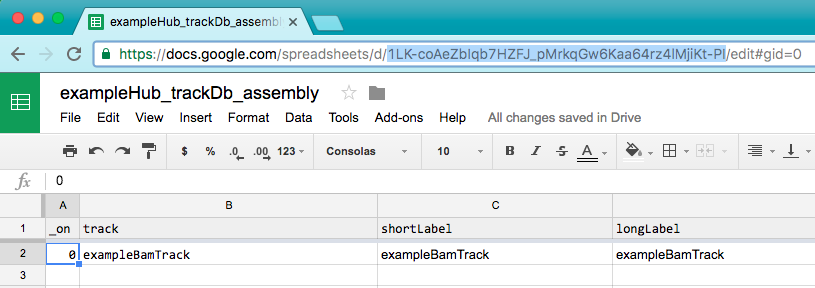
In the trackDb spreadsheet, copy the url key from the URL (hightlighted below).
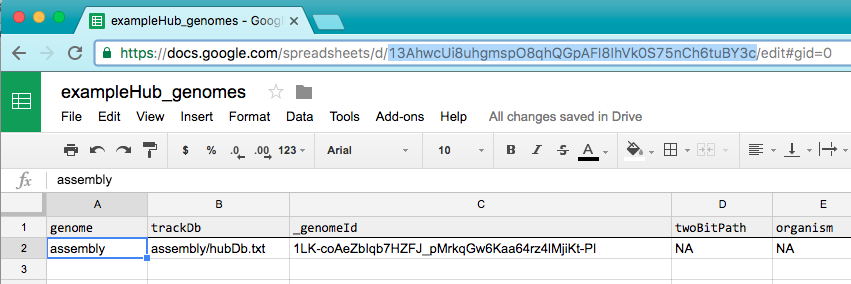
Then paste that gid into the "genome" field in the genomes spreadsheet. Then copy the url key of the genomes spreadsheet (highlighted below).
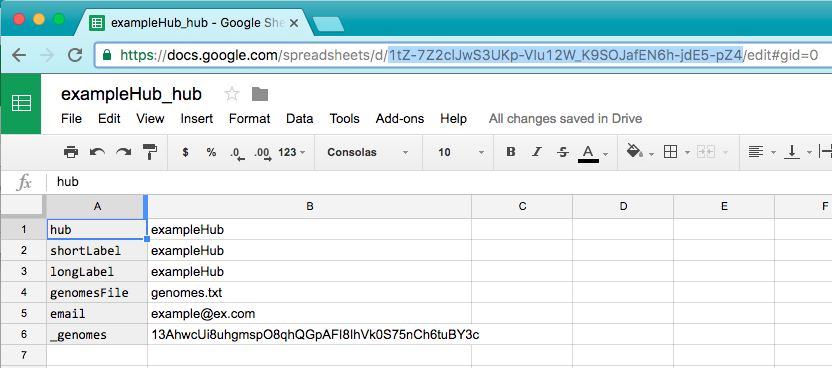
Then paste that gid into the "_genomes" field in the hub spreadsheet. Then copy the url key of the hub spreadsheet (highlighted below).
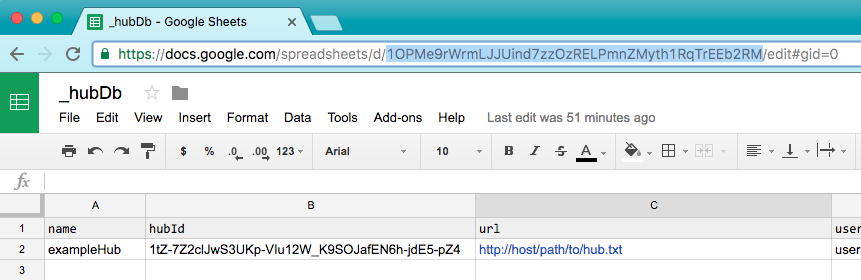
Then paste that gid into the "hubId" field in the _hubDb spreadsheet. Then copy the url key of the hubDb spreadsheet (highlighted below).
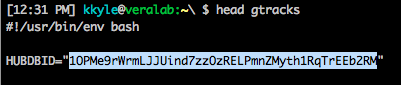
Then use a text editor to replace the key defining HUBDBID in the gtracks script with what you copied from the hubDb spreadsheet URL (highlighted below).
This process establishes a relationship between the gtracks script, the spreadsheet defining what hubs to manage, the genomes for each hub, and the tracks for each genome:
gtracks -> _hubDb -> name_hub -> name_genomes -> name_trackDb_assembly
In the directory where you saved gtracks, run "./gtracks all" to have gtracks set up the necesary directory hierarchy for the hubs defined in _hubDb.
Place big data files in the bbi directories on webserver as described above.
Go to genome.ucsc.edu -> My Data -> Track Hubs -> My Hubs and paste full url pointing to hub.txt on the webserver in URL box.
- automate generation and sharing of google spreadsheets
- error handling
- more docs
- allow an entire hub to be defined in multiple sheets within the same google spreadsheet
- create cloudinit to help users quickly spin up a web server to host hubs