Pure Java implementation of Van Der Maaten and Hinton's t-SNE clustering algorithm.
T-SNE-Java supports Barnes Hut which makes it possible to run the amazing t-SNE on much larger data sets (or much faster on small data sets:) )!
The Barnes Hut version can also be run in parallel! We have seen from 40 % performance improvements on moderate datasets (ca 10 000 samples) to 100 % improvements on larger datasets (MNIST 60000 samples) compared to standard Barnes Hut.
The t-SNE part of running the parallel Barnes Hut t-SNE on MNIST 60000 takes 18.3 minutes on a 2013 Macbook Pro (theta=0.5, perplexity = 50, 1000 iterations)
Both standard and parallel Barnes Hut is of course magnitudes faster than vanilla t-SNE.
Great research by Dr. Maaten!!
This project is divided into two separate Maven projects, one for the core t-SNE and one for the demos (stand-alone executables that can be run from command line).
If you just want to use TSne as a command line tool, you should use BarnesHutTSneCsv for the Barnes Hut version or TSneCsv for the classic version.
You must then first build and install 'tsne' and 'tsne-demos' (mvn install). Then use the tsne-demos JAR you just build according to the examples below.
You can also download the pre-build binary JAR from the release page.
Examples:
Run TSne on file without headers and no labels.
java -jar target/tsne-demos-2.4.0.jar -nohdr -nolbls src/main/resources/datasets/iris_X.txt Run TSne on CSV file with headers and label column nr. 5.
java -jar target/tsne-demos-2.4.0.jar --lblcolno 5 src/main/resources/datasets/iris.csvRun TSne on file without headers and no labels but supply a separate label file (with the same ordering as the data file).
java -jar target/tsne-demos-2.4.0.jar --nohdr --nolbls --label_file=src/main/resources/datasets/iris_X_labels.txt src/main/resources/datasets/iris_X.txtSame as above but using parallelization.
java -jar target/tsne-demos-2.4.0.jar --parallel --nohdr --nolbls --label_file=src/main/resources/datasets/iris_X_labels.txt src/main/resources/datasets/iris_X.txtThe BarnesHutTSneCsv program now supports aborting gracefully.
If the output is monitored and it is concluded that the process has converged, the BarnesHutTSneCsv process can be stopped with a graceful exit by sending the process an interrupt signal.
kill -2 <PID>The program now exits and produces the same output as usual except for the plot which must be done manually.
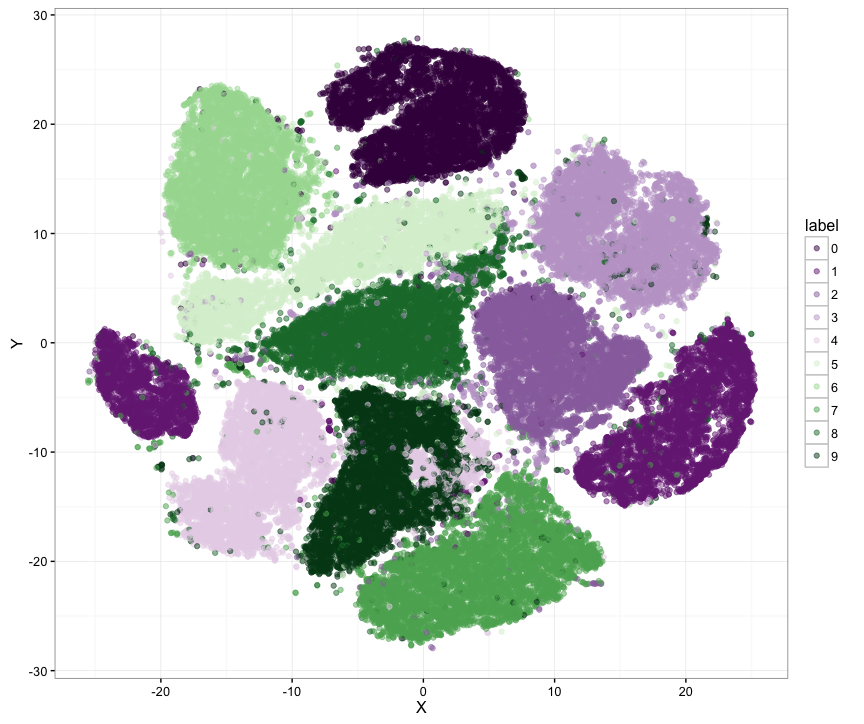
Example graph of the MNIST data set (60000 samples) generated with Barnes Hut implementation of t-SNE:
For some tips working with t-sne [Klick here] (http://lejon.github.io) or [here] (https://lvdmaaten.github.io/tsne/#faq) (observe that the last link discusses some implementation details of Laurens implementation of t-SNE and not this Java version, but also some general tips and tricks which applies to t-SNE in general) .
import java.io.File;
import com.jujutsu.tsne.barneshut.BHTSne;
import com.jujutsu.tsne.barneshut.BarnesHutTSne;
import com.jujutsu.tsne.barneshut.ParallelBHTsne;
import com.jujutsu.utils.MatrixOps;
import com.jujutsu.utils.MatrixUtils;
import com.jujutsu.utils.TSneUtils;
public class TSneTest {
public static void main(String [] args) {
int initial_dims = 55;
double perplexity = 20.0;
double [][] X = MatrixUtils.simpleRead2DMatrix(new File("src/main/resources/datasets/mnist2500_X.txt"), " ");
System.out.println(MatrixOps.doubleArrayToPrintString(X, ", ", 50,10));
BarnesHutTSne tsne;
boolean parallel = false;
if(parallel) {
tsne = new ParallelBHTsne();
} else {
tsne = new BHTSne();
}
TSneConfiguration config = TSneUtils.buildConfig(X, 2, initial_dims, perplexity, 1000);
double [][] Y = tsne.tsne(config);
// Plot Y or save Y to file and plot with some other tool such as for instance R
}
}T-SNE-Java is not on Maven Central, however you can use it through Jitpack by adding the following lines to you POM file.
<repositories>
<repository>
<id>jitpack.io</id>
<url>https://jitpack.io</url>
</repository>
</repositories>
<dependency>
<groupId>com.github.User</groupId>
<artifactId>Repo</artifactId>
<version>Tag</version>
</dependency>Demo: 2.4.0 Core: 2.5.0
I'm a very satisfied user of the YourKit profiler. A Great product with great support. It has been sucessfully used for profiling in this project.
YourKit supports open source projects with its full-featured Java Profiler. YourKit, LLC is the creator of YourKit Java Profiler and YourKit .NET Profiler, innovative and intelligent tools for profiling Java and .NET applications.
Enjoy! -Leif