-
Notifications
You must be signed in to change notification settings - Fork 2
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
This reconciles the two divergent histories of the old `brainglobe-scripts` and `cellfinder` (CLI tool).
- Loading branch information
Showing
20 changed files
with
1,553 additions
and
140 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,150 +1,139 @@ | ||
| [](https://pypi.org/project/cellfinder) | ||
| [](https://pypi.org/project/cellfinder) | ||
| [](https://pepy.tech/project/cellfinder) | ||
| [](https://pypi.org/project/cellfinder) | ||
| [](https://github.com/brainglobe/cellfinder) | ||
| []( | ||
| https://github.com/brainglobe/cellfinder/actions) | ||
| [](https://codecov.io/gh/brainglobe/cellfinder) | ||
| [](https://pypi.org/project/brainglobe-workflows) | ||
| [](https://pypi.org/project/brainglobe-workflows) | ||
| [](https://pepy.tech/project/brainglobe-workflows) | ||
| [](https://pypi.org/project/brainglobe-workflows) | ||
| [](https://github.com/brainglobe/brainglobe-workflows) | ||
| []( | ||
| https://github.com/brainglobe/brainglobe-workflows/actions) | ||
| [](https://codecov.io/gh/brainglobe/brainglobe-workflows) | ||
| [](https://github.com/python/black) | ||
| [](https://pycqa.github.io/isort/) | ||
| [](https://github.com/pre-commit/pre-commit) | ||
| [](https://docs.brainglobe.info/cellfinder/contributing) | ||
| [](https://brainglobe.info/documentation/cellfinder/index.html) | ||
| [](https://brainglobe.info/developers/index.html) | ||
| [](https://brainglobe.info/documentation/brainglobe-workflows/index.html) | ||
| [](https://twitter.com/brain_globe) | ||
|
|
||
| # BrainGlobe Workflows | ||
|
|
||
| `brainglobe-workflows` is a package that provides users with a number of out-of-the-box data analysis workflows employed in neuroscience, implemented using BrainGlobe tools. | ||
|
|
||
| At present, the package currently offers the following workflows: | ||
|
|
||
| - [cellfinder](#cellfinder): Whole-brain detection, registration, and analysis. The successor to the old [cellfinder CLI](TODO:permalnk to deprecated cellfinder tag on repo) TODO: rename tool appropriately and give flavour text | ||
|
|
||
| ## Installation | ||
|
|
||
| `brainglobe-workflows` comes packaged with version 1 of BrainGlobe, so the easiest way to make sure you get the latest release and stay up to date is to install that package - [follow this link to see the install instructions](TODO: link me!). | ||
|
|
||
| If you want to install BrainGlobe workflows as a standalone tool, you can run `pip install` in your desired environment: | ||
|
|
||
| ```bash | ||
| pip install brainglobe-workflows | ||
| ``` | ||
|
|
||
| ## Contributing | ||
|
|
||
| Contributions to BrainGlobe are more than welcome. | ||
| Please see the [developers guide](https://brainglobe.info/developers/index.html). | ||
|
|
||
| ## Citing `brainglobe-workflows` | ||
|
|
||
| **If you use any tools in the [brainglobe suite](https://brainglobe.info/documentation/index.html), please [let us know](mailto:[email protected]?subject=cellfinder), and we'd be happy to promote your paper/talk etc.** | ||
|
|
||
| If you find [`cellfinder`](#cellfinder) useful, and use it in your research, please cite the paper outlining the cell detection algorithm: | ||
| > Tyson, A. L., Rousseau, C. V., Niedworok, C. J., Keshavarzi, S., Tsitoura, C., Cossell, L., Strom, M. and Margrie, T. W. (2021) “A deep learning algorithm for 3D cell detection in whole mouse brain image datasets’ PLOS Computational Biology, 17(5), e1009074 | ||
| [https://doi.org/10.1371/journal.pcbi.1009074](https://doi.org/10.1371/journal.pcbi.1009074) | ||
| > | ||
| If you use any of the image registration functions in `cellfinder`, please also cite [`brainreg`](https://github.com/brainglobe/brainreg#citing-brainreg). | ||
|
|
||
| --- | ||
|
|
||
| # Cellfinder | ||
|
|
||
| **TODO: move this information to an appropriate place on the website** | ||
|
|
||
| Whole-brain cell detection, registration and analysis. | ||
|
|
||
| **N.B. If you want to just use the cell detection part of cellfinder, please | ||
| see the standalone [cellfinder-core](https://github.com/brainglobe/cellfinder-core) | ||
| package, or the [cellfinder plugin](https://github.com/brainglobe/cellfinder-napari) | ||
| for [napari](https://napari.org/).** | ||
| **N.B. If you want to just use the cell detection part of cellfinder, please see the standalone [cellfinder-core](https://github.com/brainglobe/cellfinder-core) package, or the [cellfinder plugin](https://github.com/brainglobe/cellfinder-napari) for [napari](https://napari.org/).** | ||
|
|
||
| --- | ||
| `cellfinder` is a collection of tools developed by [Adam Tyson](https://github.com/adamltyson), [Charly Rousseau](https://github.com/crousseau) and [Christian Niedworok](https://github.com/cniedwor) in the [Margrie Lab](https://www.sainsburywellcome.org/web/groups/margrie-lab), generously supported by the [Sainsbury Wellcome Centre](https://www.sainsburywellcome.org/web/). | ||
|
|
||
| `cellfinder` is a designed for the analysis of whole-brain imaging data such as | ||
| [serial-section imaging](https://sainsburywellcomecentre.github.io/OpenSerialSection/) | ||
| and lightsheet imaging in cleared tissue. The aim is to provide a single solution for: | ||
|
|
||
| * Cell detection (initial cell candidate detection and refinement using | ||
| deep learning) (using [cellfinder-core](https://github.com/brainglobe/cellfinder-core)) | ||
| * Atlas registration (using [brainreg](https://github.com/brainglobe/brainreg)) | ||
| * Analysis of cell positions in a common space | ||
| `cellfinder` is a designed for the analysis of whole-brain imaging data such as [serial-section imaging](https://sainsburywellcomecentre.github.io/OpenSerialSection/) and lightsheet imaging in cleared tissue. | ||
| The aim is to provide a single solution for: | ||
|
|
||
| --- | ||
| Installation is with | ||
| `pip install cellfinder` | ||
| - Cell detection (initial cell candidate detection and refinement using deep learning) (using [cellfinder-core](https://github.com/brainglobe/cellfinder-core)), | ||
| - Atlas registration (using [brainreg](https://github.com/brainglobe/brainreg)), | ||
| - Analysis of cell positions in a common space. | ||
|
|
||
| --- | ||
| Basic usage: | ||
|
|
||
| ```bash | ||
| cellfinder -s signal_images -b background_images -o output_dir --metadata metadata | ||
| ``` | ||
| Full documentation can be | ||
| found [here](https://brainglobe.info/documentation/cellfinder/index.html). | ||
|
|
||
| This software is at a very early stage, and was written with our data in mind. | ||
| Over time we hope to support other data types/formats. If you have any issues, please get in touch [on the forum](https://forum.image.sc/tag/brainglobe) or by | ||
| [raising an issue](https://github.com/brainglobe/cellfinder/issues/new/choose). | ||
| Full documentation can be found [here](https://brainglobe.info/documentation/cellfinder/index.html). | ||
|
|
||
| This software is at a very early stage, and was written with our data in mind. | ||
| Over time we hope to support other data types/formats. | ||
| If you have any issues, please get in touch [on the forum](https://forum.image.sc/tag/brainglobe) or by [raising an issue](https://github.com/brainglobe/cellfinder/issues/new/choose). | ||
|
|
||
| --- | ||
| ## Illustration | ||
|
|
||
| ### Introduction | ||
| cellfinder takes a stitched, but otherwise raw whole-brain dataset with at least | ||
| two channels: | ||
| * Background channel (i.e. autofluorescence) | ||
| * Signal channel, the one with the cells to be detected: | ||
|
|
||
|  | ||
| **Raw coronal serial two-photon mouse brain image showing labelled cells** | ||
| cellfinder takes a stitched, but otherwise raw whole-brain dataset with at least two channels: | ||
|
|
||
| - Background channel (i.e. autofluorescence), | ||
| - Signal channel, the one with the cells to be detected: | ||
|
|
||
|  | ||
|
|
||
| ### Cell candidate detection | ||
| Classical image analysis (e.g. filters, thresholding) is used to find | ||
| cell-like objects (with false positives): | ||
|
|
||
|  | ||
| **Candidate cells (including many artefacts)** | ||
| Classical image analysis (e.g. filters, thresholding) is used to find cell-like objects (with false positives): | ||
|
|
||
|  | ||
|
|
||
| ### Cell candidate classification | ||
| A deep-learning network (ResNet) is used to classify cell candidates as true | ||
| cells or artefacts: | ||
|
|
||
|  | ||
| **Cassified cell candidates. Yellow - cells, Blue - artefacts** | ||
| A deep-learning network (ResNet) is used to classify cell candidates as true cells (yellow) or artefacts (blue): | ||
|
|
||
|  | ||
|
|
||
| ### Registration and segmentation (brainreg) | ||
| Using [brainreg](https://github.com/brainglobe/brainreg), | ||
| cellfinder aligns a template brain and atlas annotations (e.g. | ||
| the Allen Reference Atlas, ARA) to the sample allowing detected cells to be assigned | ||
| a brain region. | ||
| ### Registration and segmentation (`brainreg`) | ||
|
|
||
| This transformation can be inverted, allowing detected cells to be | ||
| transformed to a standard anatomical space. | ||
| Using [`brainreg`](https://github.com/brainglobe/brainreg), `cellfinder` aligns a template brain and atlas annotations (e.g. the Allen Reference Atlas, ARA) to the sample allowing detected cells to be assigned a brain region. | ||
|
|
||
|  | ||
| **ARA overlaid on sample image** | ||
| This transformation can be inverted, allowing detected cells to be transformed to a standard anatomical space. | ||
|
|
||
|  | ||
|
|
||
| ### Analysis of cell positions in a common anatomical space | ||
| Registration to a template allows for powerful group-level analysis of cellular | ||
| disributions. *(Example to come)* | ||
|
|
||
| Registration to a template allows for powerful group-level analysis of cellular distributions. | ||
| *(Example to come)* | ||
|
|
||
| ## Examples | ||
|
|
||
| *(more to come)* | ||
|
|
||
| ### Tracing of inputs to retrosplenial cortex (RSP) | ||
| Input cell somas detected by cellfinder, aligned to the Allen Reference Atlas, | ||
| and visualised in [brainrender](https://github.com/brainglobe/brainrender) along | ||
|
|
||
| Input cell somas detected by cellfinder, aligned to the Allen Reference Atlas, and visualised in [brainrender](https://github.com/brainglobe/brainrender) along | ||
| with RSP. | ||
|
|
||
|  | ||
|
|
||
| Data courtesy of Sepiedeh Keshavarzi and Chryssanthi Tsitoura. [Details here](https://www.youtube.com/watch?v=pMHP0o-KsoQ) | ||
| Data courtesy of Sepiedeh Keshavarzi and Chryssanthi Tsitoura. | ||
| [Details here](https://www.youtube.com/watch?v=pMHP0o-KsoQ) | ||
|
|
||
| ## Visualisation | ||
|
|
||
| cellfinder comes with a plugin ([brainglobe-napari-io](https://github.com/brainglobe/brainglobe-napari-io)) for [napari](https://github.com/napari/napari) to view your data | ||
|
|
||
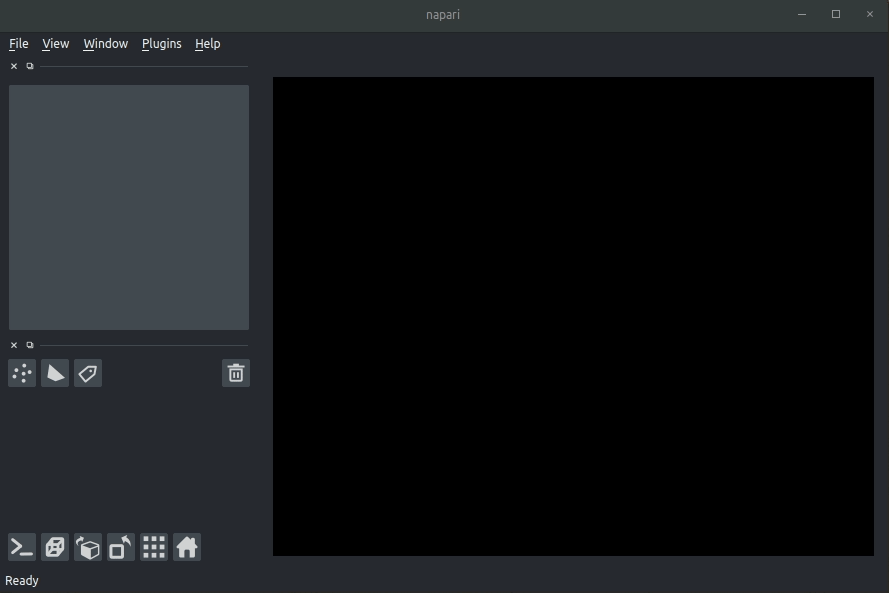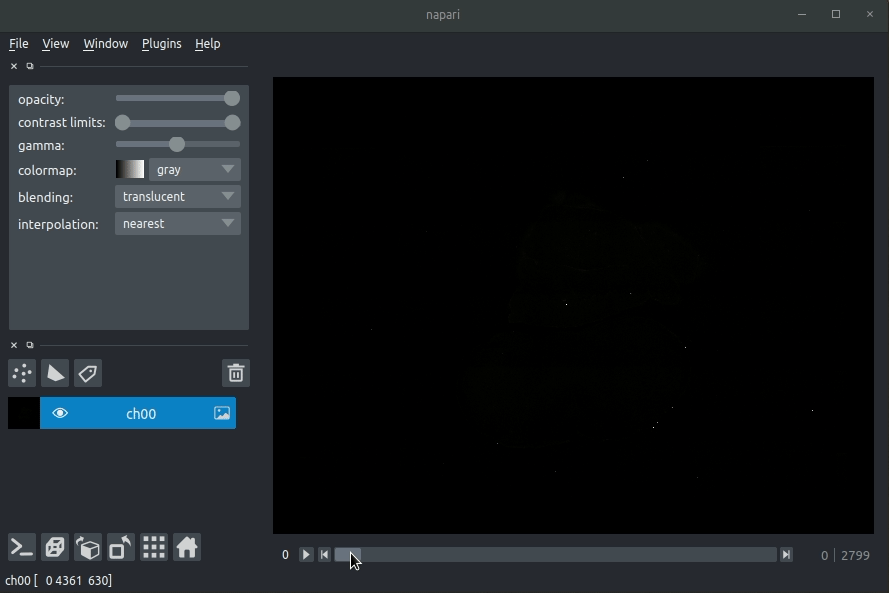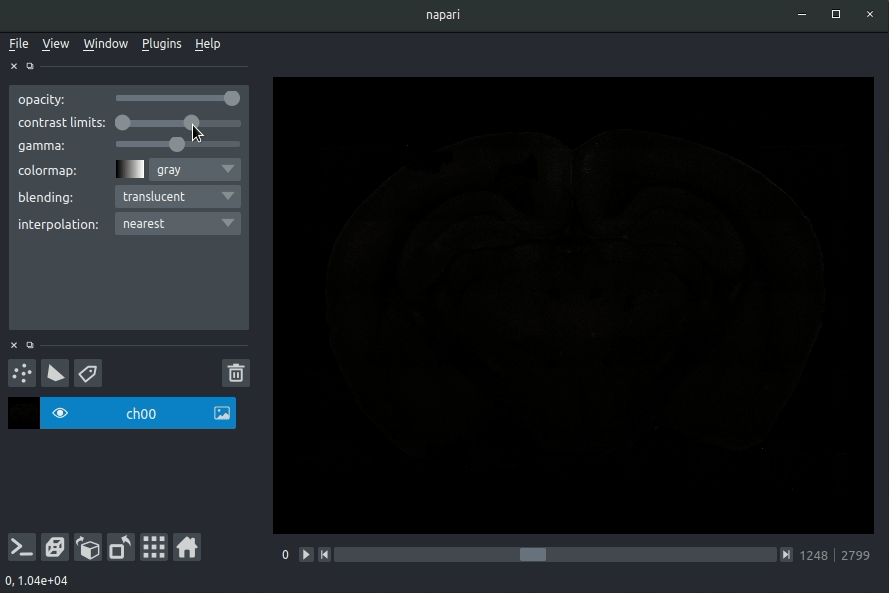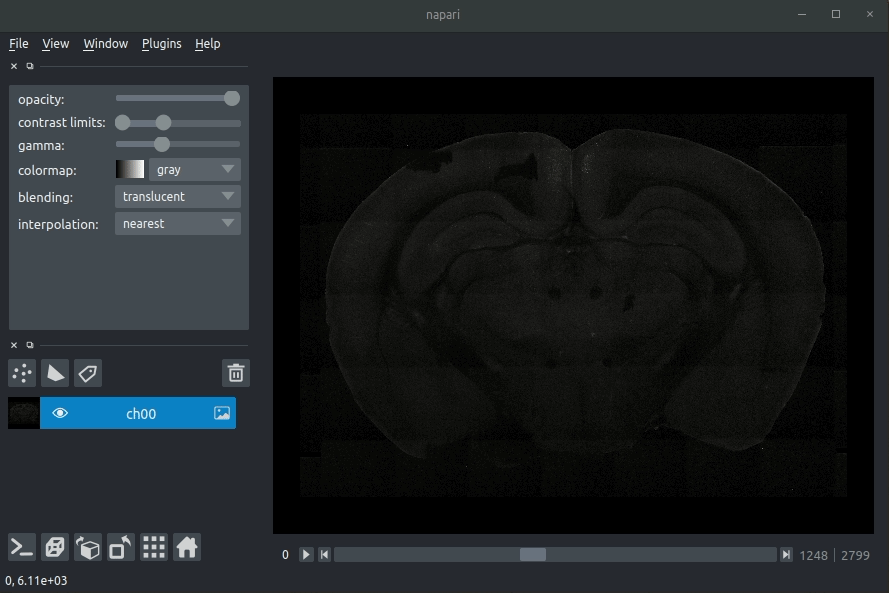
| #### Usage | ||
| * Open napari (however you normally do it, but typically just type `napari` into your terminal, or click on your desktop icon) | ||
|
|
||
| #### Load cellfinder XML file | ||
| * Load your raw data (drag and drop the data directories into napari, one at a time) | ||
| * Drag and drop your cellfinder XML file (e.g. `cell_classification.xml`) into napari. | ||
| You can view your data using the [brainglobe-napari-io](https://github.com/brainglobe/brainglobe-napari-io) plugin for [napari](https://github.com/napari/napari). | ||
|
|
||
| #### Load cellfinder directory | ||
| * Load your raw data (drag and drop the data directories into napari, one at a time) | ||
| * Drag and drop your cellfinder output directory into napari. | ||
|
|
||
| The plugin will then load your detected cells (in yellow) and the rejected cell | ||
| candidates (in blue). If you carried out registration, then these results will be | ||
| overlaid (similarly to the loading brainreg data, but transformed to the | ||
| coordinate space of your raw data). | ||
|
|
||
|  | ||
| **Loading raw data** | ||
|
|
||
| 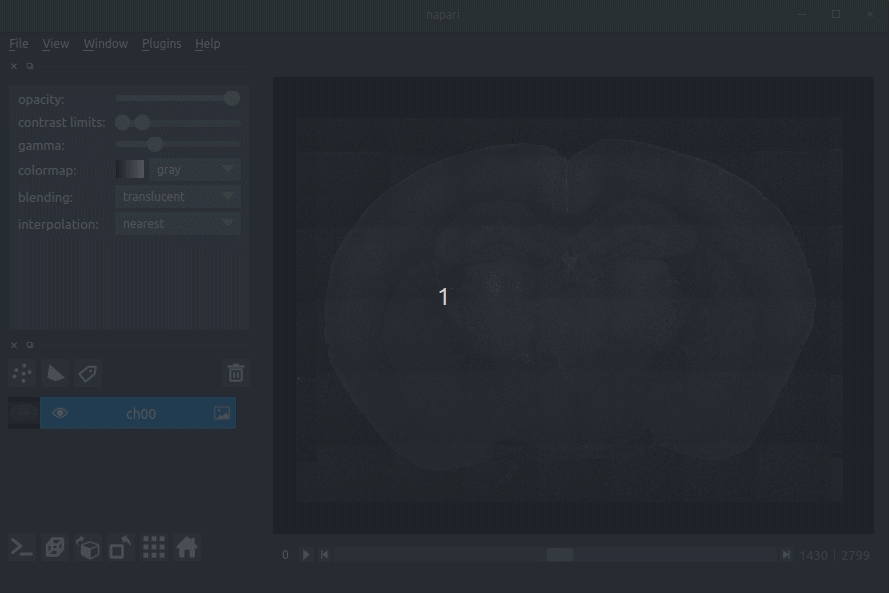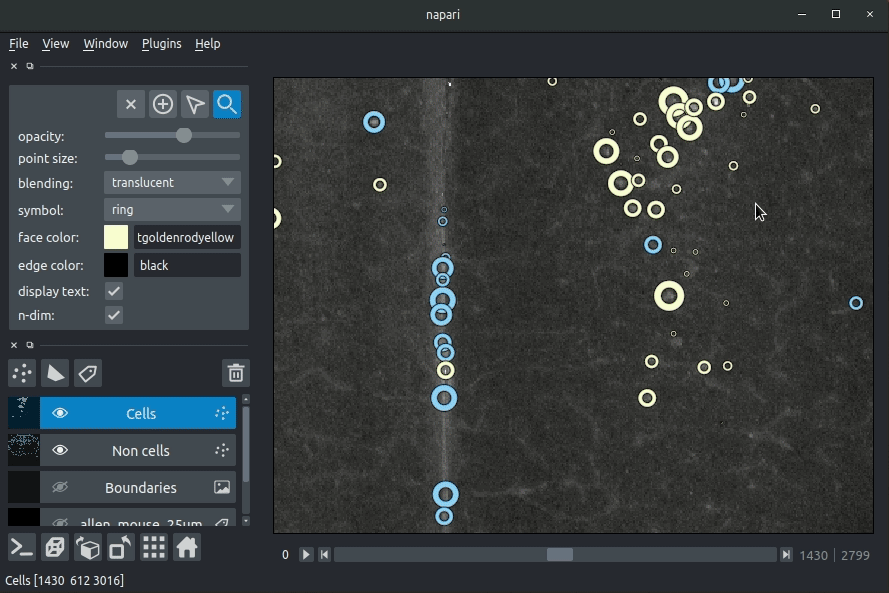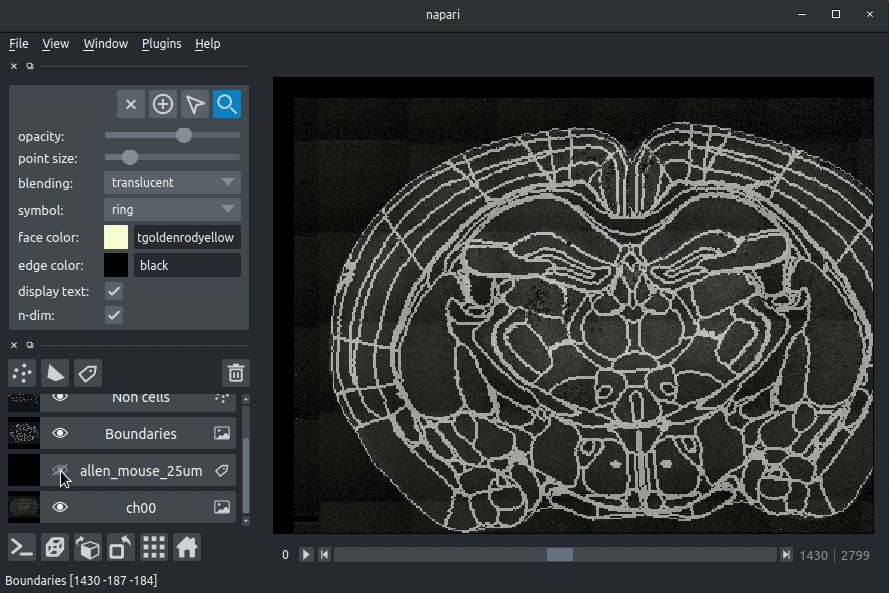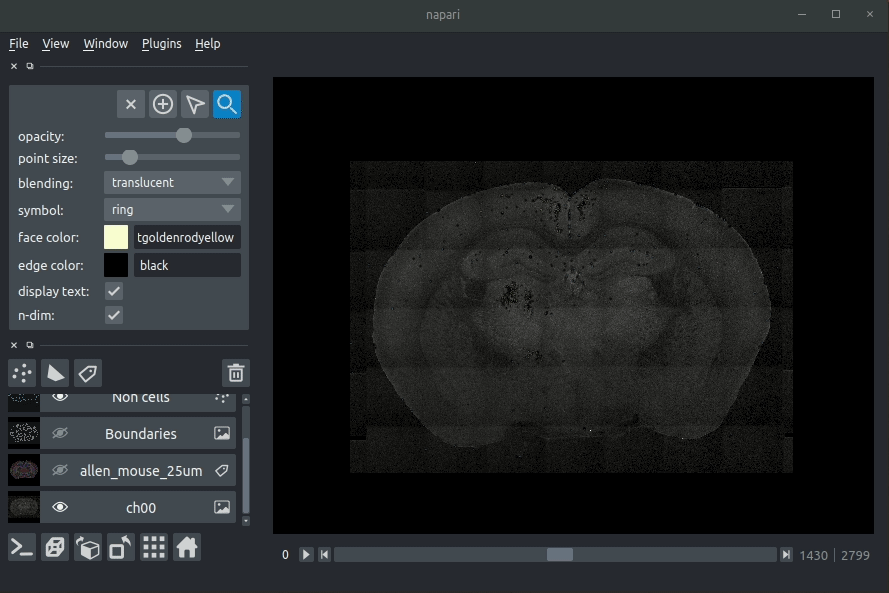 | ||
| **Loading cellfinder results** | ||
|
|
||
|
|
||
| ## Contributing | ||
| Contributions to cellfinder are more than welcome. Please see the [developers guide](https://brainglobe.info/developers/index.html). | ||
|
|
||
|
|
||
| ## Citing cellfinder | ||
|
|
||
| If you find cellfinder useful, and use it in your research, please cite the paper outlining the cell detection algorithm: | ||
| > Tyson, A. L., Rousseau, C. V., Niedworok, C. J., Keshavarzi, S., Tsitoura, C., Cossell, L., Strom, M. and Margrie, T. W. (2021) “A deep learning algorithm for 3D cell detection in whole mouse brain image datasets’ PLOS Computational Biology, 17(5), e1009074 | ||
| [https://doi.org/10.1371/journal.pcbi.1009074](https://doi.org/10.1371/journal.pcbi.1009074) | ||
| > | ||
| If you use any of the image registration functions in cellfinder, please also cite [brainreg](https://github.com/brainglobe/brainreg#citing-brainreg). | ||
| - Open napari (however you normally do it, but typically just type `napari` into your terminal, or click on your desktop icon). | ||
| - Load your raw data (drag and drop the data directories into napari, one at a time).  | ||
| - Drag and drop your cellfinder XML file (e.g. `cell_classification.xml`) and/or cellfinder output directory into napari.  | ||
|
|
||
| **If you use this, or any other tools in the brainglobe suite, please | ||
| [let us know](mailto:[email protected]?subject=cellfinder), and | ||
| we'd be happy to promote your paper/talk etc.** | ||
| The plugin will then load your detected cells (in yellow) and the rejected cell candidates (in blue). | ||
| If you carried out registration, then these results will be overlaid (similarly to the loading `brainreg` data, but transformed to the coordinate space of your raw data). |
Oops, something went wrong.