An Agent-Based Model of Monocyte Differentiation into Tumor-Associated Macrophages in Chronic Lymphocytic Leukemia
This repository contains data related to the publication An Agent-Based Model of Monocyte Differentiation into Tumor-Associated Macrophages in Chronic Lymphocytic Leukemia by Verstraete et al.
It presents an agent-based model (ABM) describing the differentiation dynamics of monocytes into tumour-associated macrophages upon contact with cancer B cells in the context of chronic lymphocytic leukemia. The goal of this ABM is to reproduce dynamics compatible with an in vitro experimental setting of monocytes differentiation in presence of leukemic (B-CLL).
The model can be run online at https://www.netlogoweb.org/. To do this:
- download the NetLogo model here (
right clicktheRawbutton at the top of the file, selectSave Link As…, choose the location on your computer where you want to save the file, and selectSave). - go at https://www.netlogoweb.org/ and click on NetLogo Web
- upload the model
- click
setupandgo.
- netlogo source file for the agent-based model.
- openmole source files for exploring the model on the OpenMOLE platform https://openmole.org
- figures exploration outputs and source files used to generate the figures and analysis of the related publication
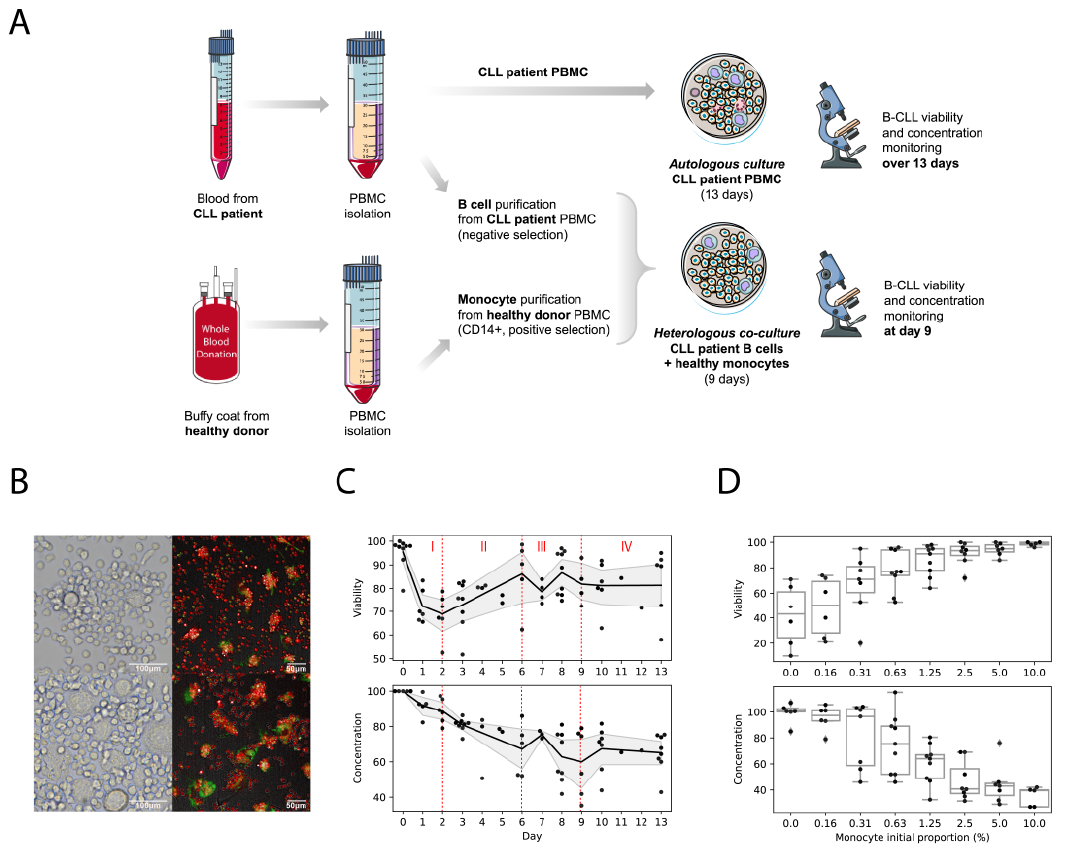 Fig.1 Experimental setups and datasets from in vitro PBMC cultures from CLL patients. A) Experimental set-up. B) Visualization of NLCs at 10 days of in vitro culture from two different patients. C) Time course datasets produced from the PBMC autologous cultures from 9 patients. D) Heterologous co-cultures.
Fig.1 Experimental setups and datasets from in vitro PBMC cultures from CLL patients. A) Experimental set-up. B) Visualization of NLCs at 10 days of in vitro culture from two different patients. C) Time course datasets produced from the PBMC autologous cultures from 9 patients. D) Heterologous co-cultures.
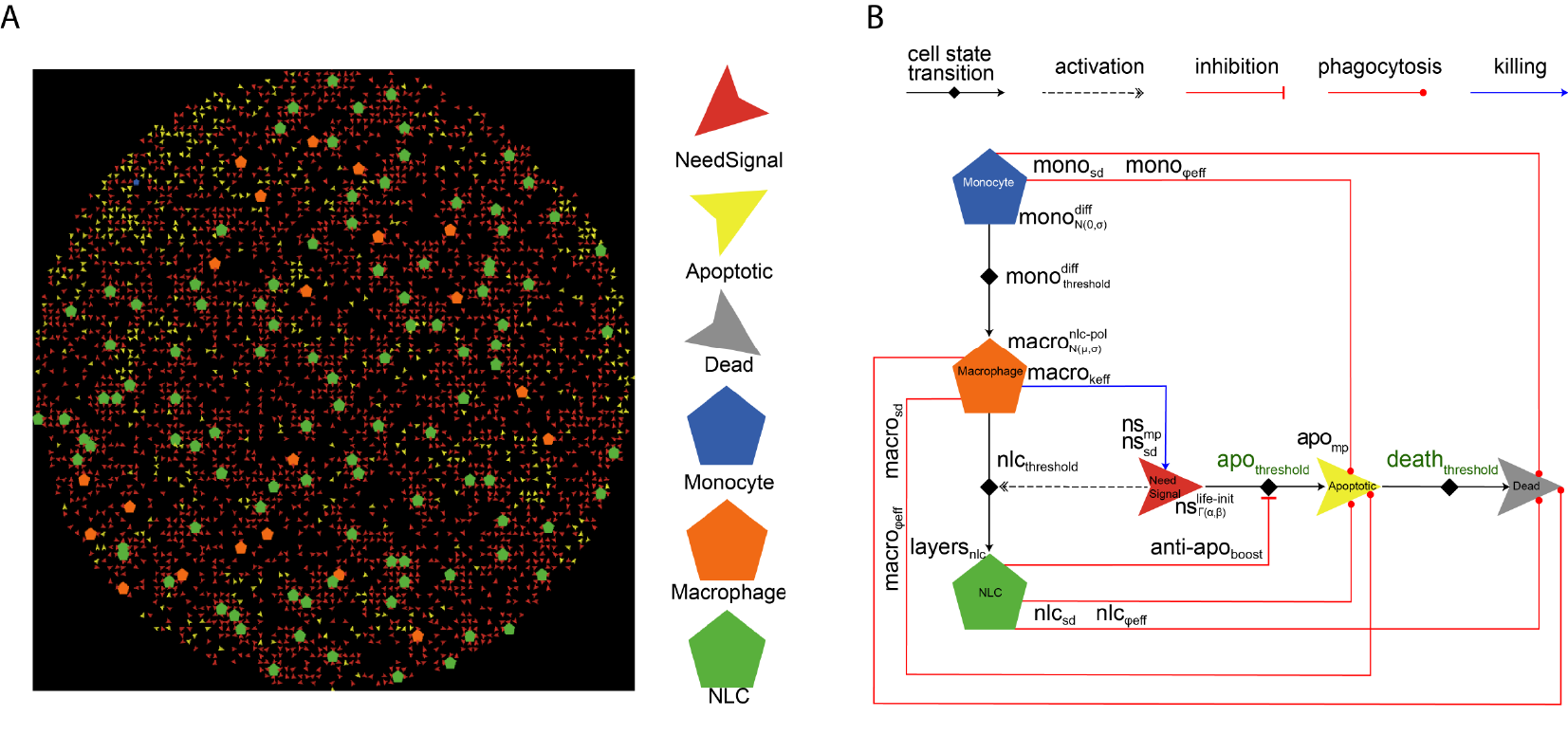 Fig. 2. ABM representations. A) Netlogo simulation of 5000 cells. B) Schematic diagram of the agents’ states and behaviors.
Fig. 2. ABM representations. A) Netlogo simulation of 5000 cells. B) Schematic diagram of the agents’ states and behaviors.
- A video of a simulation of 5000 cells can be found at https://doi.org/10.6084/m9.figshare.21592368.v1.
- A video of a live-imaging microscopy capture attempting to acquire cell pattern formations is available at https://doi.org/10.6084/m9.figshare.22110926.v1.