-
Notifications
You must be signed in to change notification settings - Fork 1
Home
Welcome to the ucvm_plotting wiki!
-
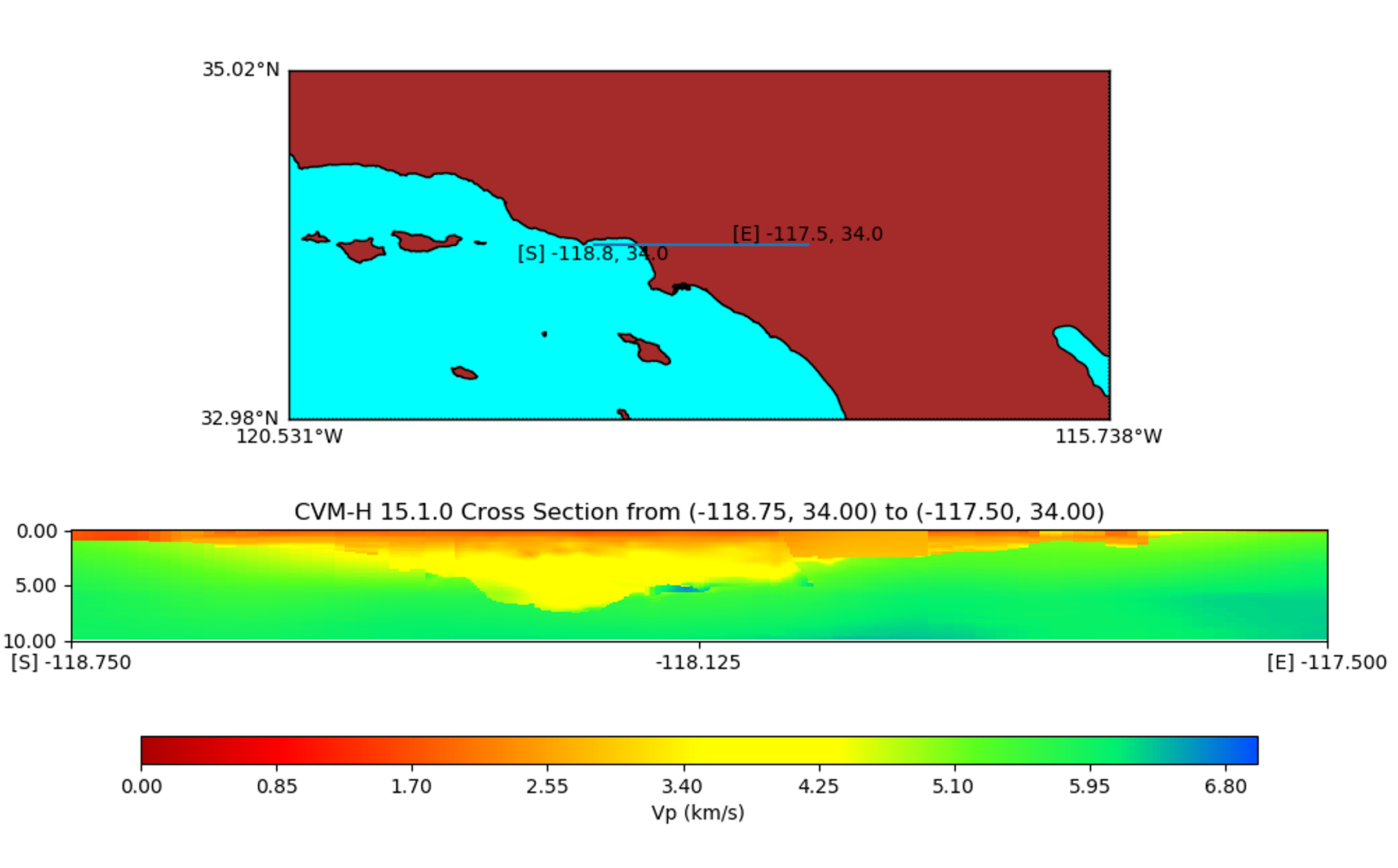
make_plots.py - A test script that generates a series of cross section plots for each supported model and one horizontal slice for cvmh
-
make_bicolor_plots.py - A test script that create elevation cross sections and cross sections plots and also horizontal plots with different gating threshold to showcase the bicolor color scale
-
make_density_plots.py - A test script that generates grid with material properties and make density plots for different model
-
make_norcal_plots.py - A test script that creates horizontal map plots of varying depth from 0m to 4000m with cencal,cca,cvmsi
-
make_scatter_plots.py - A test script that generates grid with material properties and make scatter plots for different models
This script produces a plot for each cvm:
./plot_horizontal_slice.py -b 30.5,-126.0 -u 42.5,-112.5 -s 0.05 -e 0.0 -d poisson -a s -c cs173h -o cs173h_poisson_map.png
./plot_cross_section.py -b 34.0,-122.00 -u 34.0,-117.5 -s 0 -e 2000 -h 500 -v 10 -d vs -a d -c cvmh -o cross-cvmh.png
./plot_cross_section.py -b 34.0,-122.00 -u 34.0,-117.5 -s 0 -e 2000 -h 500 -v 10 -d vs -a d -c cvms -o cross-cvms.png
./plot_cross_section.py -b 34.0,-122.00 -u 34.0,-117.5 -s 0 -e 2000 -h 500 -v 10 -d vs -a d -c cvmsi -o cross-cvmsi.png
./plot_cross_section.py -b 34.0,-122.00 -u 34.0,-117.5 -s 0 -e 2000 -h 500 -v 10 -d vs -a d -c cvms5 -o cross-cvms5.png
./plot_cross_section.py -b 34.0,-122.00 -u 34.0,-117.5 -s 0 -e 2000 -h 500 -v 10 -d vs -a d -c 1d -o cross-1d.png
./plot_cross_section.py -b 34.0,-122.00 -u 34.0,-117.5 -s 0 -e 2000 -h 500 -v 10 -d vs -a d -c bbp1d -o cross-bbp1d.png
./plot_cross_section.py -b 34.0,-122.00 -u 34.0,-117.5 -s 0 -e 2000 -h 500 -v 10 -d vs -a d -c cencal -o cross-cencal.png
./plot_cross_section.py -b 34.0,-122.00 -u 34.0,-117.5 -s 0 -e 2000 -h 500 -v 10 -d vs -a d -c cca -o cross-cca.png
./plot_horizontal_slice.py -b 33.5,-118.75 -u 34.5,-117.5 -s 0.1 -e 500 -d vs -a d -c cvmh -o horizontal-cvmh.png
./plot_horizontal_slice.py -b 33.5,-118.75 -u 34.5,-117.5 -s 0.1 -e 500 -d vs -a d -c cvms -o horizontal-cvms.png
./plot_horizontal_slice.py -b 33.5,-118.75 -u 34.5,-117.5 -s 0.1 -e 500 -d vs -a d -c cvmsi -o horizontal-cvmsi.png
./plot_horizontal_slice.py -b 33.5,-118.75 -u 34.5,-117.5 -s 0.1 -e 500 -d vs -a d -c cvms5 -o horizontal-cvms5.png
./plot_horizontal_slice.py -b 33.5,-118.75 -u 34.5,-117.5 -s 0.1 -e 500 -d vs -a d -c 1d -o horizontal-1d.png
./plot_horizontal_slice.py -b 33.5,-118.75 -u 34.5,-117.5 -s 0.1 -e 500 -d vs -a d -c bbp1d -o horizontal-bbp1d.png
./plot_horizontal_slice.py -b 33.5,-118.75 -u 34.5,-117.5 -s 0.1 -e 500 -d vs -a d -c cencal -o horizontal-cencal.png
./plot_horizontal_slice.py -b 33.5,-118.75 -u 34.5,-117.5 -s 0.1 -e 500 -d vs -a d -c cca -o horizontal-cca.png
The following commands will produce plots in bi-color colormap with user selected gating point (default is 2500m)
./plot_cross_section.py -b 35.21,-121 -u 35.21,-118 -c cca -s 0 -e 5000 -v 100 -h 1000 -g 2.5 -a b -d vs -o cross_section_cca_bi.png
./plot_elevation_cross_section.py -b 35.21,-121 -u 35.21,-118 -h 1000 -v -100 -d vs -c cca -a b -s 1000 -e -4000 -g 2.5 -o cross_section_elevation_cca_b.png
./plot_cross_section.py -c cvmh -s 0 -e 5000 -v 100 -h 1000 -g 2.5 -a b -d vs -b 35.21,-121 -u 35.21,-118 -o cross_section_cvmh_bi.png
./plot_horizontal_slice.py -b 34.21,-121.0 -u 36.21,-118 -s 0.05 -e 200 -d vs -a b -c cca -o horizontal_cca_vs_bi.png






















