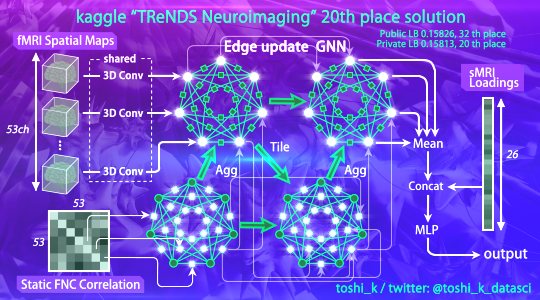
My solution in this Kaggle competition "TReNDS Neuroimaging", 20th place.
Channel-wise 3D convolutions are applied to fMRI spatial maps. All channels share the weight of convolution to prevent overfitting. Output features are thrown into Edge Update GNN with FNC correlation.
Subsequently the outputs form GNN are averaged and concatenated with sMRI loading. Finally, conventional MLP is applied and prediction for age and other target variables are obtained.
- numpy==1.14.0
- pandas==0.25.3
- scikit-learn==0.21.3
- nilearn==0.6.2
- chainer==7.4.0
- h5py==2.8.0
The preprocess function for fMRI spatial maps is forked from Rohit's public notebook.
- TReNDS - EDA + Visualization + Simple Baseline
https://www.kaggle.com/rohitsingh9990/trends-eda-visualization-simple-baseline
The custom loss function to optimize normalized absolute errors is forked from Tawara's public notebook.
- TReNDS:Simple NN Baseline
https://www.kaggle.com/ttahara/trends-simple-nn-baseline
- Neural Message Passing with Edge Updates for Predicting Properties of Molecules and Materials
Peter Bjørn Jørgensen, Karsten Wedel Jacobsen, Mikkel N. Schmidt
https://arxiv.org/abs/1806.03146