LIBSVM Accelerated with GPU using the CUDA Framework
GPU-accelerated LIBSVM is a modification of the original LIBSVM that exploits the CUDA framework to significantly reduce processing time while producing identical results. The functionality and interface of LIBSVM remains the same. The modifications were done in the kernel computation, that is now performed using the GPU.
Watch a short video on the capabilities of the GPU-accelerated LIBSVM package
###CHANGELOG
V1.2
Updated to LIBSVM version 3.17
Updated to CUDA SDK v5.5
Using CUBLAS_V2 which is compatible with the CUDA SDK v4.0 and up.
Mode Supported
C-SVC classification with RBF kernel
Functionality / User interface
Same as LIBSVM
LIBSVM prerequisites
NVIDIA Graphics card with CUDA support
Latest NVIDIA drivers for GPU
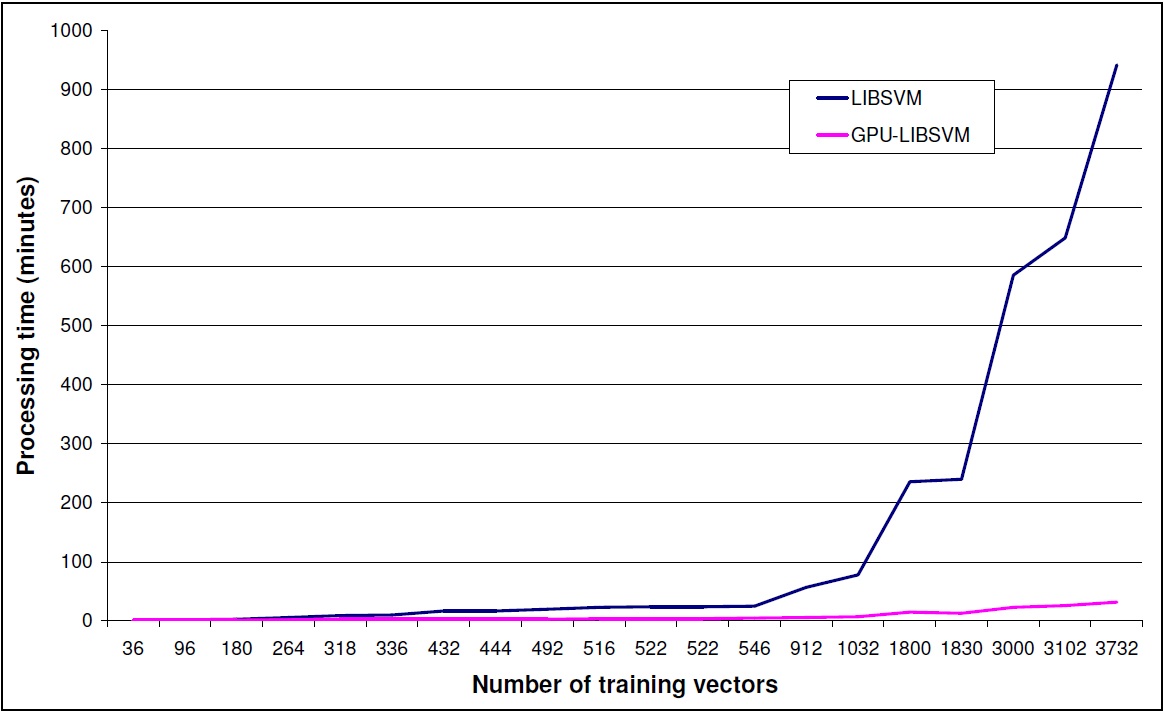
To showcase the performance gain using the GPU-accelerated LIBSVM we present an example run.
PC Setup
Quad core Intel Q6600 processor
3.5GB of DDR2 RAM
Windows-XP 32-bit OS
Input Data
TRECVID 2007 Dataset for the detection of high level features in video shots
Training vectors with a dimension of 6000
20 different feature models with a variable number of input training vectors ranging from 36 up to 3772
Classification parameters
c-svc
RBF kernel
Parameter optimization using the easy.py script provided by LIBSVM.
4 local workers
Discussion
GPU-accelerated LIBSVM gives a performance gain depending on the size of input data set.
This gain is increasing dramatically with the size of the dataset.
Please take into consideration input data size limitations that can occur from the memory
capacity of the graphics card that is used.
A first document describing some of the work related to the GPU-Accelerated LIBSVM is the following; please cite it if you find this implementation useful in your work:
A. Athanasopoulos, A. Dimou, V. Mezaris, I. Kompatsiaris, "GPU Acceleration for Support Vector Machines", Proc. 12th International Workshop on Image Analysis for Multimedia Interactive Services (WIAMIS 2011), Delft, The Netherlands, April 2011.
THIS SOFTWARE IS PROVIDED BY THE AUTHOR "AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE AUTHOR BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
- Is there a GPU-accelerated LIBSVM version for Matlab?
We are interested in porting our imlementation in Matlab but due to our workload, it is not in our immediate plans. Everybody is welcome to make the port and we can host the ported software.
- Visual Studio will not let me build the provided project.
The project has been built in both 32/64 bit mode. If you are working on 32 bits, you might need the x64 compiler for Visual Studio 2010 to build the project.
- Building the project, I get linker error messages.
Please go to the project properties and check the library settings. CUDA libraries have different names for x32 / x64. Make sure that the correct path and filenames are given.
- I have built the project but the executables will not run (The application has failed to start because its side-by-side configuration is in correct.)
Please update the VS2010 redistributables to the PC you are running your executable and install all the latest patches for visual studio.
- My GPU-accelerated LIBSVM is running smoothly but i do not see any speed-up.
GPU-accelerated LIBSVM is giving speed-ups mainly for big datasets. In the GPU-accelerated implementation some extra time is needed to load the data to the gpu memory. If the dataset is not big enough to give a significant performance gain, the gain is lost due to the gpu-memory -> cpu-memory, cpu-memory -> gpu-memory transfer time. Please refer to the graph above to have a better understanding of the performance gain for different dataset sizes. Problems also seem to arise when the input dataset contains values with extreme differences (e.g. 107) if no scaling is performed. Such an example is the "breast-cancer" dataset provided in the official LIBSVM page.
This work was supported by the EU FP7 projects GLOCAL (FP7-248984) and WeKnowIt (FP7-215453)