💫 Graph-based foundation model for spatial transcriptomics data
Novae is a deep learning model for spatial domain assignments of spatial transcriptomics data (at both single-cell or spot resolution). It works across multiple gene panels, tissues, and technologies. Novae offers several additional features, including: (i) native batch-effect correction, (ii) analysis of spatially variable genes and pathways, and (iii) architecture analysis of tissue slides.
Check Novae's documentation to get started. It contains installation explanations, API details, and tutorials.
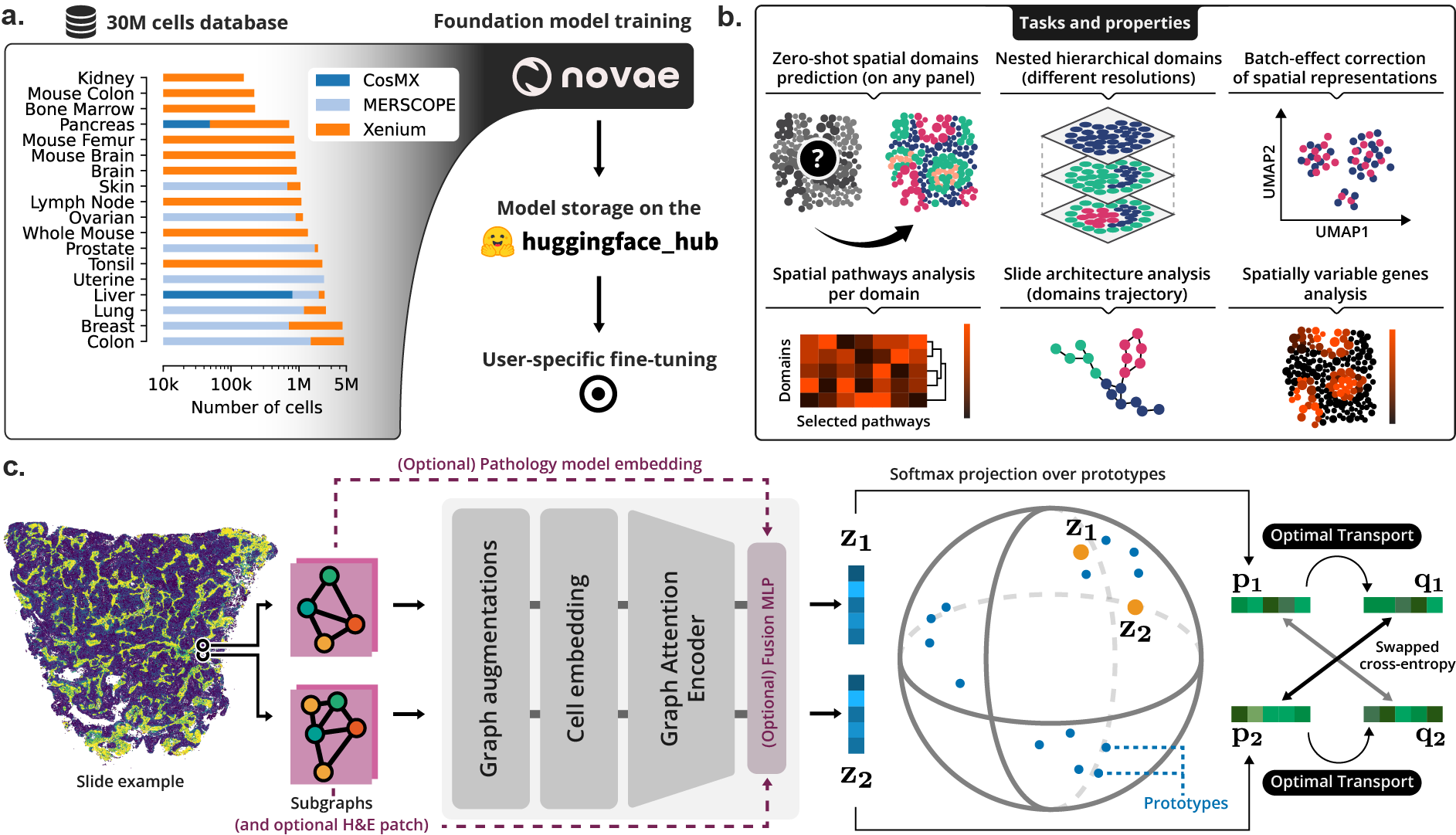
(a) Novae was trained on a large dataset, and is shared on Hugging Face Hub. (b) Illustration of the main tasks and properties of Novae. (c) Illustration of the method behing Novae (self-supervision on graphs, adapted from SwAV).
novae can be installed via PyPI on all OS, for python>=3.9.
pip install novae
To install novae in editable mode (e.g., to contribute), clone the repository and choose among the options below.
pip install -e . # pip, minimal dependencies
pip install -e '.[dev]' # pip, all extras
poetry install # poetry, minimal dependencies
poetry install --all-extras # poetry, all extrasHere is a minimal usage example. For more details, refer to the documentation.
import novae
model = novae.Novae.from_pretrained("MICS-Lab/novae-human-0")
model.compute_representations(adata, zero_shot=True)
model.assign_domains(adata)You can cite our preprint as below:
@article{blampeyNovae2024,
title = {Novae: A Graph-Based Foundation Model for Spatial Transcriptomics Data},
author = {Blampey, Quentin and Benkirane, Hakim and Bercovici, Nadege and Andre, Fabrice and Cournede, Paul-Henry},
year = {2024},
pages = {2024.09.09.612009},
publisher = {bioRxiv},
doi = {10.1101/2024.09.09.612009},
}