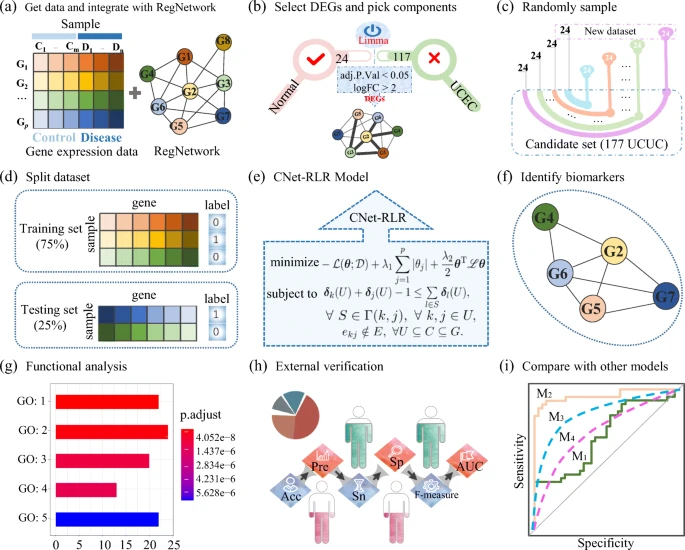
In this work, a connected network-regularized logistic regression (CNet-RLR) model for feature selection considering the structural connectivity in a network was proposed. Mathematically, it was a convex optimization problem constrained by inequalities reflecting network connectivity.
- CNet: A connected network-regularized logistic regression (CNet-RLR) model were proposed to perform feature selection.
- In both synthetic simulation data and real-world UCEC cancer genomics data, we validated the CNet-RLR model is efficient to identify the connected-network-structured features that can serve as diagnostic biomarkers.
- In the comparison study, we also proved the proposed CNet-RLR model results in better classification performance and feature interpretability than the other regularized logistic regression (RLR) alternatives and another graph embedded feature selection model.
- If you have any questions about CNet, please directly contact the corresponding author Prof. Zhi-Ping Liu with the E-mail: [email protected]
Li, Lingyu, and Zhi-Ping Liu. "A connected network-regularized logistic regression model for feature selection." Applied Intelligence (2022): 1-31. CoxReg paper website
- In the CNet-RLR, NSLR_example and matlab_example files, we give all R/Matlab/Python codes used in our work.
- In the Data and GSE63678 files, we give some necessary input/output files by the R/Matlab/Python codes.
- Some of these input files only give the first few lines, but this does not affect the results of the work (CNet-RLR).
- In the Supplementary file file, we present the necessary Additional files contained in our work.
The serial number (1), (2), ..., (9) represents the order in which the program runs in our work.
- (1) network_match.R ---- Network structure of DEGs in RegNetwork
- (2) Data_net.R ---- Intersection of genes in the Data and genes in RegNetwork
- (3) DE_net.R ---- Select differentially expressed genes (DEGs) and consider the network structure in RegNetwork
- (4) data_split2.R ---- Randomly divide Dataset into 20 new Datasets by 1:1 of the positive and negative samples
- (5) cut.R ---- Diameters and cut-nodes of component of DEGs in RegNetwork
- (6) adj.R ---- Adjacency matrix and its eigenvalues
- (7) CNet_RLR_126e50102.m ---- Main function
- (8) coef2feature_data ---- Select feature genes based on threshold and Extract feature expression values in test dataset and independent dataset
- (9) class_net.R ---- Independent data set verification to obtain ROC curve and classification accuracy
- costFunction12.m ---- Objective function
- cv.m ---- Cross validation to select optimal parameters
- Laplcian_Matrix.m ---- Laplacian matrix according to the adjacency matrix
- LogitisLap.m ---- LogitisLap function for cv
- SGNLR.m ---- SGNLR function for LogitisLap
- my_error.m ---- error function
- getLambMax.m ---- getLambMax function for cv
- Predict.m ---- Predict function on test dataset
- plot_roc.m ---- Roc curve function on test dataset
- RunRcode.m ---- RunRcode for main function
This program package is supported by the copyright owners and coders "as is" and without warranty of any kind, express or implied, including, but not limited to, the implied warranties of merchantability and fitness for a particular purpose. In no event shall the copyright owner or contributor be liable for any direct, indirect, incidental, special, exemplary, or consequential damages (including, without limitation, procurement of substitute goods or services; loss of use, data, or profits; or business interruption), regardless of the theory of liability, whether in contract, strict liability or tort (including negligence or otherwise) for any use of the software, even if advised of the possibility of such damages.