NeuroPyxels (npyx) is a python library built for electrophysiologists using Neuropixels electrodes. This package stems from the need of a pythonist who really did not want to transition to MATLAB to work with Neuropixels: it features a suite of core utility functions for loading, processing and plotting Neuropixels data.
There isn't any better doc atm - post an issue if you have any question, or email Maxime Beau (PhD Hausser lab, UCL). You can also use the Neuropixels slack workgroup.
- Documentation
- Load synchronyzation channel
- Get good units from dataset
- Load spike times from unit u
- Load waveforms from unit u
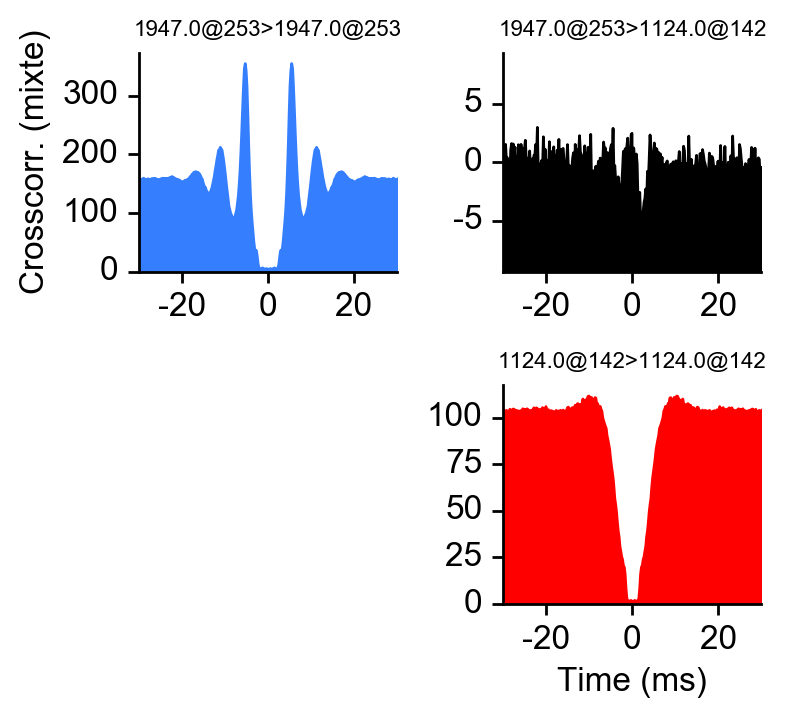
- Compute auto/crosscorrelogram between 2 units
- Plot waveform and crosscorrelograms of unit u
- Preprocess your waveforms and spike trains
- Plot chunk of raw data with overlaid units
- Plot peri-stimulus time histograms across neurons and conditions
- Merge datasets acquired on two probes simultaneously
- Bonus: cool npyx plotting utilities which could turn out useful
- Installation
- Acknowledgement
- Developer cheatsheet
Npyx works in harmony with the data formatting employed by SpikeGLX and OpenEphys used in combination with Kilosort or SpyKING CIRCUS and Phy ([after conversion for the phy gui](https://spyking-circus.readthedocs.io/en/latest/GUI/launching.html, results stored in path/mydata/mydata.GUI)). Any dataset you can run phy on can also be analyzed with npyx, in essence.
Npyx is fast because it never computes the same thing twice - in the background, it saves most relevant outputs (spike trains, waveforms, correlograms...) at kilosort_dataset/npyxMemory, from where they are simply reloaded if called again. An important parameter controlling this behaviour is again (boolean), by default set to False: if True, the function will recompute the output rather than loading it from npyxMemory. This is important to be aware of this behaviour, as it can lead to mind boggling bugs. For instance, if you load the train of unit then re-spikesort your dataset, e.g. you split unit 56 in 504 and 505, the train of the old unit 56 will still exist at kilosort_dataset/npyxMemory and you will be able to load it even though the unit is gone!
Most npyx functions take at least one input: dp, which is the path to your Kilosort-phy dataset. You can find a full description of the structure of such datasets on phy documentation.
Other typical parameters are: verbose (whether to print a bunch of informative messages, useful when debugging), saveFig (boolean) and saveDir (whether to save the figure in saveDir for plotting functions).
Importantly, dp can also be the path to a merged dataset, generated with npyx.merge_datasets() - every function will run as smoothly on merged datasets as on any regular dataset. See below for more details.
More precisely, every function requires the files myrecording.ap.meta/myrecording.oebin (metadata from SpikeGLX/OpenEphys), params.py, spike_times.npy and spike_clusters.npy. If you have started spike sorting, cluster_groups.tsv will also be required obviously (will be created filled with 'unsorted' groups if none is found). Then particular functions will require particular files: loading waveforms with npyx.spk_wvf.wvf or extracting your sync channel with npyx.io.get_npix_sync require the raw data myrecording.ap.bin``myrecording.dat, npyx.spk_wvf.templates the files templates.npy and spike_templates.npy, and so on. This allows you to only transfer the strictly necassary files for your use case from a machine to the next: for instance, if you only want to make behavioural analysis of spike trains but do not care about the waveforms, you can run get_npix_sync on a first machine (which will generate a sync_chan folder containing extracted onsets/offsets from the sync channel(s)), then exclusively transfer the sync_chan folder along with spike_times.npy and spike_clusters.npy (all very light files) on another computer and analyze your data there seemlessly.
DISCLAIMER: some parts of the code (in particular contents of feat.py) are experimental - use at your own risk!
The dp parameter of all npyx functions must be the absolute path to myrecording below.
For SpikeGLX recordings:
myrecording/
myrecording.ap.meta
params.py
spike_times.npy
spike_clusters.npy
cluster_groups.tsv # optional, if manually curated with phy
myrecording.ap.bin # optional, if wanna plot waveforms
# other kilosort/spyking circus outputs here
For Open-Ephys recordings:
myrecording/
myrecording.oebin
params.py
spike_times.npy
spike_clusters.npy
cluster_groups.tsv # if manually curated with phy
# other kilosort/spyking circus outputs here
continuous/
Neuropix-PXI-100.0/
continuous.dat # optional, if wanna plot waveforms
Neuropix-PXI-100.1/
continuous.dat # optional, if want to plot LFP with plot_raw
events/
Neuropix-PXI-100.0/
TTL_1/
timestamps.npy # optional, if need to get synchronyzation channel to load with get_npix_sync e.g. to merge datasets
Example use cases are:
from npyx import *
dp = 'datapath/to/myrecording'
# load contents of .lf.meta and .ap.meta or .oebin files as python dictionnary.
# The metadata of the high and lowpass filtered files are in meta['highpass'] and meta['lowpass']
# Quite handy to get probe version, sampling frequency, recording length etc
meta = read_metadata(dp)from npyx.inout import get_npix_sync # star import is sufficient, but I like explicit imports!
# If SpikeGLX: slow the first time, then super fast
onsets, offsets = get_npix_sync(dp)
# onsets/offsets are dictionnaries
# keys: ids of sync channel where a TTL was detected,
# values: times of up (onsets) or down (offsets) threshold crosses, in seconds.Makes a preprocessed copy of the binary file in dp, moves original binary file at dp/original_data This will be as fast as literally copying your file, with a decent GPU!
from npyx.inout import preprocess_binary_file # star import is sufficient, but I like explicit imports!
# can perform bandpass filtering (butterworth 3 nodes) and median subtraction (aka common average referenceing, CAR)
# in the future: ADC realignment (like CatGT), whitening, spatial filtering (experimental).
filtered_fname = preprocess_binary_file(dp, filt_key='ap', median_subtract=True, f_low=None, f_high=300, order=3, verbose=True)from npyx.gl import get_units
units = get_units(dp, quality='good')from npyx.spk_t import trn
u=234
t = trn(dp, u) # gets all spikes from unit 234, in samplesfrom npyx.inout import read_spikeglx_meta
from npyx.spk_t import ids, trn
from npyx.spk_wvf import get_peak_chan, wvf, templates
# returns a random sample of 100 waveforms from unit 234, in uV, across 384 channels
waveforms = wvf(dp, u) # return array of shape (n_waves, n_samples, n_channels)=(100, 82, 384) by default
waveforms = wvf(dp, u, n_waveforms=1000, t_waveforms=90) # now 1000 random waveforms, 90 samples=3ms long
# Get the unit peak channel (channel with the biggest amplitude)
peak_chan = get_peak_chan(dp,u)
# extract the waveforms located on peak channel
w=waves[:,:,peak_chan]
# Extract waveforms of spikes occurring between
# 0-100s and 300-400s in the recording,
# because that's when your mouse sneezed
waveforms = wvf(dp, u, periods=[(0,100),(300,400)])
# alternatively, longer but more flexible:
fs=meta['highpass']['sampling_rate']
t=trn(dp,u)/fs # convert in s
# get ids of unit u: all spikes have a unique index in the dataset,
# which is their rank sorted by time (as in spike_times.npy)
u_ids = ids(dp,u)
ids=ids(dp,u)[(t>900)&(t<1000)]
mask = (t<100)|((t>300)&(t<400))
waves = wvf(dp, u, spike_ids=u_ids[mask])
# If you want to load the templates instead (faster and does not require binary file):
temp = templates(dp,u) # return array of shape (n_templates, 82, n_channels)from npyx.corr import ccg, ccg_stack
# returns ccg between 234 and 92 with a binsize of 0.2 and a window of 80
c = ccg(dp, [234,92], cbin=0.2, cwin=80)
# Only using spikes from the first and third minutes of recording
c = ccg(dp, [234,92], cbin=0.2, cwin=80, periods=[(0,60), (120,180)])
# better, compute a big stack of crosscorrelograms with a given name
# The first time, CCGs will be computed in parallel using all the available CPU cores
# and it will be saved in the background and, reloadable instantaneously in the future
source_units = [1,2,3,4,5]
target_units = [6,7,8,9,10]
c_stack = ccg_stack(dp, source_units, target_units, 0.2, 80, name='my_relevant_ccg_stack')
c_stack = ccg_stack(dp, name='my_relevant_ccg_stack') # will work to reaload in the future# all plotting functions return matplotlib figures
from npyx.plot import plot_wvf, get_peak_chan
u=234
# plot waveform, 2.8ms around templates center, on 16 channels around peak channel
# (the peak channel is found automatically, no need to worry about finding it)
fig = plot_wvf(dp, u, Nchannels=16, t_waveforms=2.8)
# But if you wished to get it, simply run
peakchannel = get_peak_chan(dp, u)# plot ccg between 234 and 92
# as_grid also plot the autocorrelograms
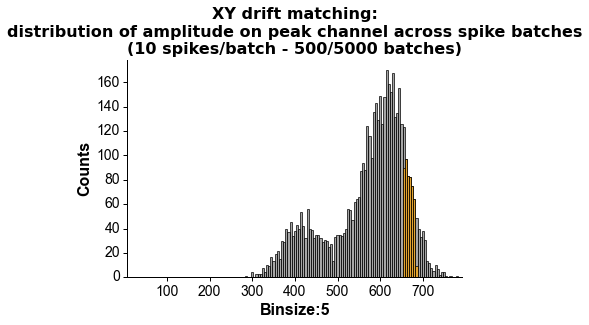
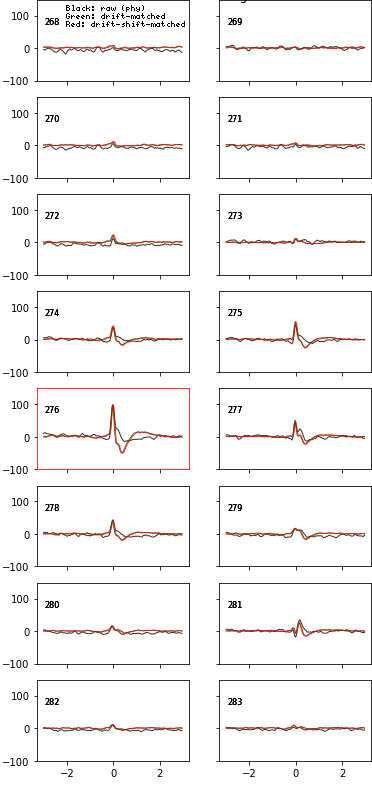
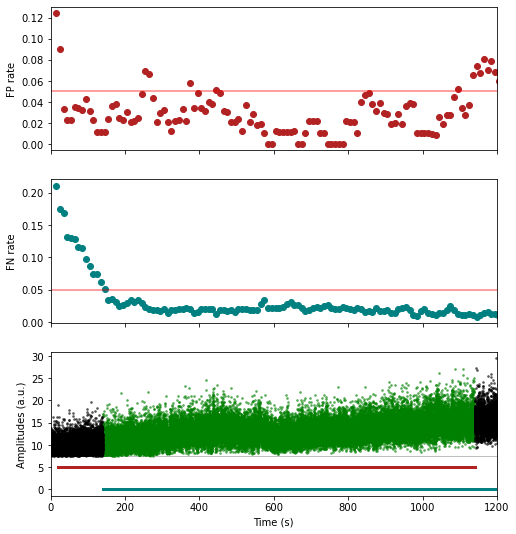
fig = plot_ccg(dp, [u,92], cbin=0.2, cwin=80, as_grid=True)Preprocess your waveforms (drift-shift-matching) and spike trains (detect periods with few false positive/negative)
# all plotting functions return matplotlib figures
from npyx.spk_wvf import wvf_dsmatch
from npyx.spk_t import trn_filtered
# wvf_dsmatch subselect 'best looking' waveforms
# by first matching them by drift state (Z, peak channel and XY, amplitude on peak channel)
# then shifting them around to realign them (using the crosscorr of its whole spatial footprint)
# on the plot, black is the original waveform as it would be plotted in phy,
# green is drift-matched, red is drift-shift matched
w_preprocessed = wvf_dsmatch(dp, u, plot_debug=True)# trn_filtered clips the recording in 10s (default) chunks
# and estimates the false positive/false negative spike sporting rates on such chunks
# before masking out spikes occurring inside 'bad chunks',
# defined as chunks with too high FP OR FN rates (5% and 5% by default)
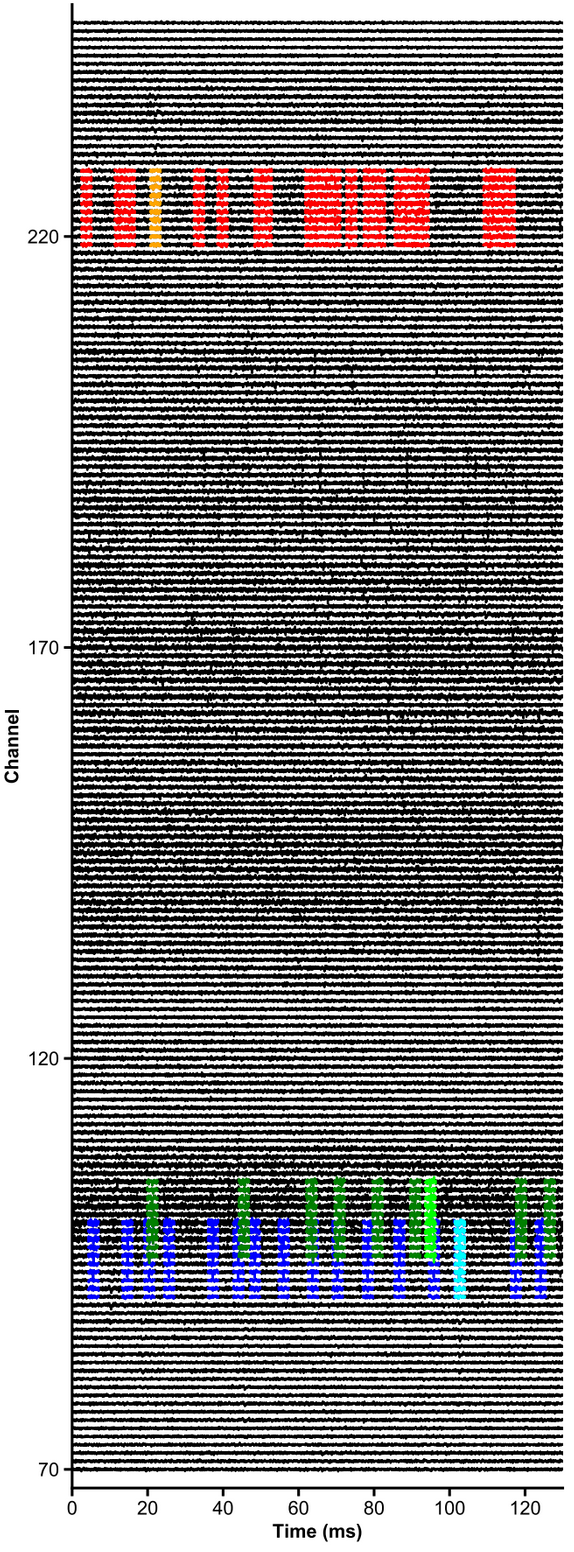
t_preprocessed = trn_filtered(dp, u, plot_debug=True)units = [1,2,3,4,5,6]
channels = np.arange(70,250)
# raw data are whitened, high-pass filtered and median-subtracted by default - parameters are explicit below
plot_raw_units(dp, times=[0,0.130], units = units, channels = channels,
colors=['orange', 'red', 'limegreen', 'darkgreen', 'cyan', 'navy'],
lw=1.5, offset=450, figsize=(6,16), Nchan_plot=10,
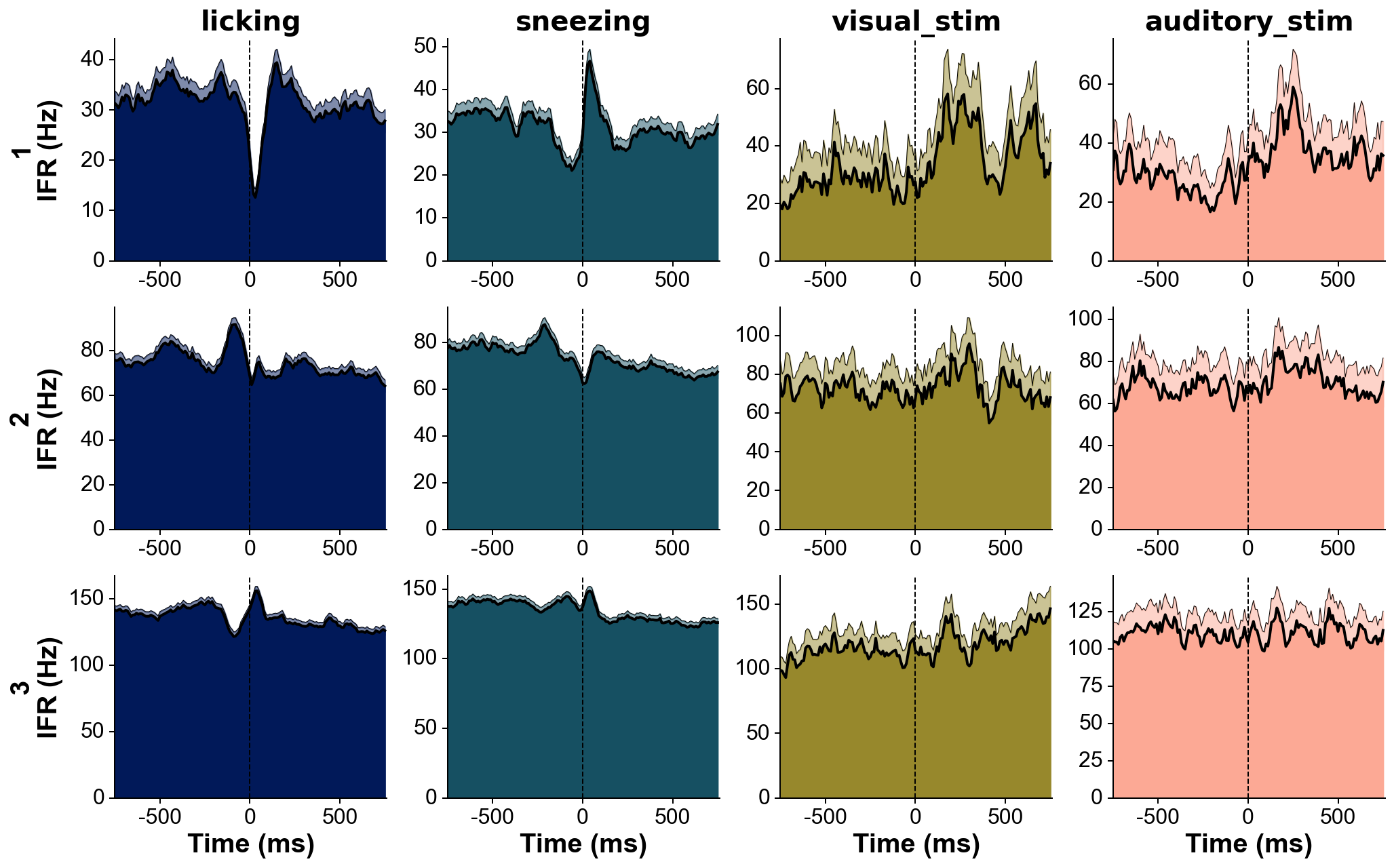
med_sub=1, whiten=1, hpfilt=1)# Explore responses of 3 neurons to 4 categories of events:
fs=30000 # Hz
units=[1,2,3]
trains=[trn(dp,u)/fs for u in units] # make list of trains of 3 units
trains_str=units # can give specific names to units here, show on the left of each row
events=[licks, sneezes, visual_stimuli, auditory_stimuli] # get events corresponding to 4 conditions
trains_str=['licking', 'sneezing', 'visual_stim', 'auditory_stim'] # can give specific names to events here, show above each column
events_col='batlow' # colormap from which the event colors will be drawn
fig=summary_psth(trains, trains_str, events, events_str, psthb=10, psthw=[-750,750],
zscore=0, bsl_subtract=False, bsl_window=[-3000,-750], convolve=True, gsd=2,
events_toplot=[0], events_col=events_col, trains_col_groups=trains_col_groups,
title=None, saveFig=0, saveDir='~/Downloads', _format='pdf',
figh=None, figratio=None, transpose=1,
as_heatmap=False, vmin=None, center=None, vmax=None, cmap_str=None)# The three recordings need to include the same sync channel.
from npyx.merger import merge_datasets
dps = ['same_folder/lateralprobe_dataset',
'same_folder/medialprobe_dataset',
'same_folder/anteriorprobe_dataset']
probenames = ['lateral','medial','anterior']
dp_dict = {p:dp for p, dp in zip(dps, probenames)}
# This will merge the 3 datasets (only relevant information, not the raw data) in a new folder at
# dp_merged: same_folder/merged_lateralprobe_dataset_medialprobe_dataset_anteriorprobe_dataset
# where all npyx functions can smoothly run.
# The only difference is that units now need to be called as floats,
# of format u.x (u=unit id, x=dataset id [0-2]).
# lateralprobe, medial probe and anteriorprobe x will be respectively 0,1 and 2.
dp_merged, datasets_table = merge_datasets(dp_dic)
--- Merged data (from 2 dataset(s)) will be saved here: /same_folder/merged_lateralprobe_dataset_medialprobe_dataset_anteriorprobe_dataset.
--- Loading spike trains of 2 datasets...
sync channel extraction directory found: /same_folder/lateralprobe_dataset/sync_chan
Data found on sync channels:
chan 2 (201 events).
chan 4 (16 events).
chan 5 (175 events).
chan 6 (28447 events).
chan 7 (93609 events).
Which channel shall be used to synchronize probes? >>> 7
sync channel extraction directory found: /same_folder/medialprobe_dataset/sync_chan
Data found on sync channels:
chan 2 (201 events).
chan 4 (16 events).
chan 5 (175 events).
chan 6 (28447 events).
chan 7 (93609 events).
Which channel shall be used to synchronize probes? >>> 7
sync channel extraction directory found: /same_folder/anteriorprobe_dataset/sync_chan
Data found on sync channels:
chan 2 (201 events).
chan 4 (16 events).
chan 5 (175 events).
chan 6 (28194 events).
chan 7 (93609 events).
Which channel shall be used to synchronize probes? >>> 7
--- Aligning spike trains of 2 datasets...
More than 50 sync signals found - for performance reasons, sub-sampling to 50 homogenoeously spaced sync signals to align data.
50 sync events used for alignement - start-end drift of -3080.633ms
--- Merged spike_times and spike_clusters saved at /same_folder/merged_lateralprobe_dataset_medialprobe_dataset_anteriorprobe_dataset.
--> Merge successful! Use a float u.x in any npyx function to call unit u from dataset x:
- u.0 for dataset lateralprobe_dataset,
- u.1 for dataset medialprobe_dataset,
- u.2 for dataset anteriorprobe_dataset.Now any npyx function runs on the merged dataset!
Under the hood, it will create a merged_dataset_dataset1_dataset2/npyxMemory folder to save any data computed across dataframes, but will use the original dataset1/npyxMemory folder to save data related to this dataset exclusively (e.g. waveforms). Hence, there is no redundancy: space and time are saved.
This is also why it is primordial that you do not move your datatasets from their original paths after merging them - else, functions ran on merged_dataset1_dataset2 will not know where to go fetch the data! They refer to the paths in merged_dataset_dataset1_dataset2/datasets_table.csv. If you really need to, you can move your datasets but do not forget to edit this file accordingly.
# These will work!
t = trn(dp_merged, 92.1) # get spikes of unit 92 in dataset 1 i.e. medialprobe
fig=plot_ccg(dp_merged,[10.0, 92.1, cbin=0.2, cwin=80]) # compute CCG between 2 units across datasetsPS - The spike times are aligned across datasets by modelling the drift between the clocks of the neuropixels headstages linearly: TTL probe 1 = a * TTL probe 1 + b (if a!=1, there is drift between the clocks), so spiketimes_probe2_aligned_to_probe1 = a * spiketimes_probe2 + b
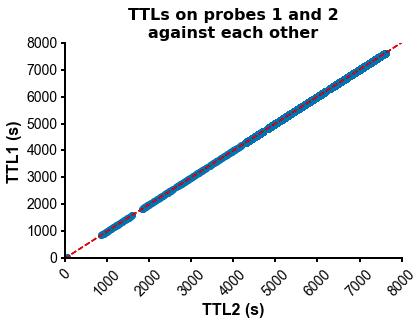
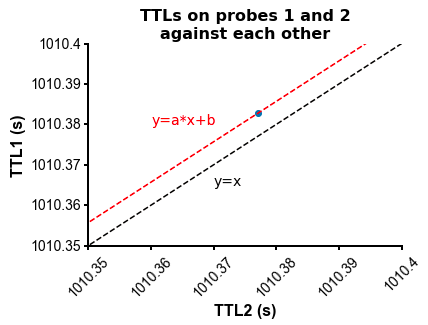
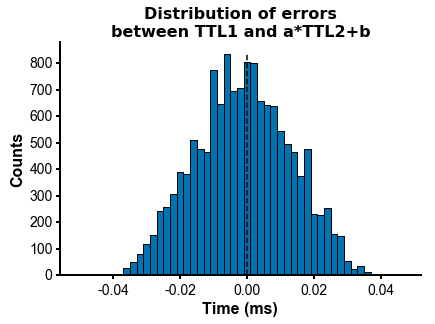
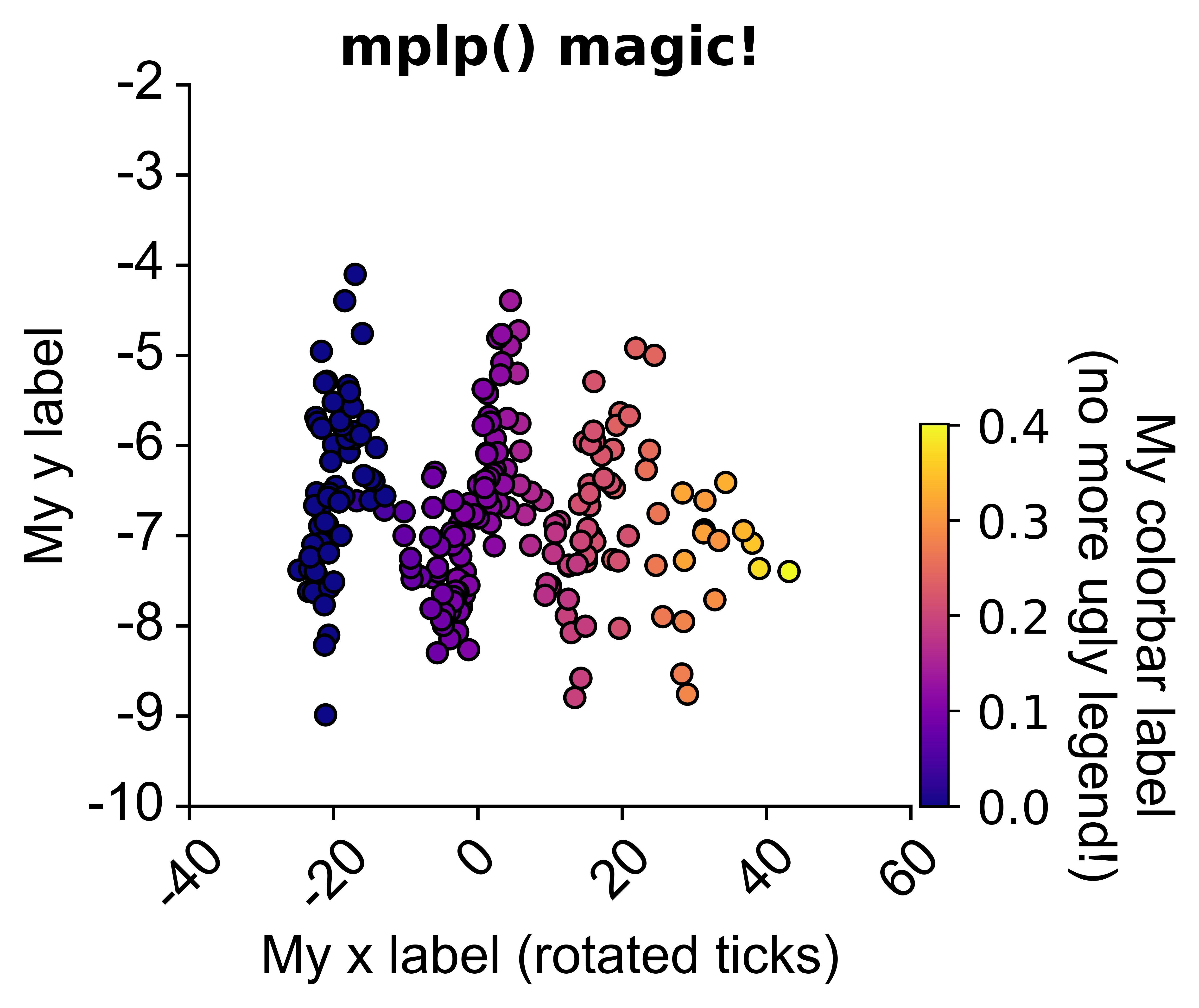
from npyx.plot import get_ncolors_cmap
# allows you to easily extract the (r,g,b) tuples from a matplotlib or crameri colormap
# to use them in other plots!
colors = get_ncolors_cmap('coolwarm', 10, plot=1)
colors = get_ncolors_cmap('viridis', 10, plot=1)
# in a jupyter notebook, will also plot he HTML colormap:from npyx.plot import mplp
import matplotlib.pyplot as plt
# mplp() will turn any matplotlib plot into something you can work with.
# fed up googling around and landing on stack overflow to tweak your figures?
# just read mplp parameters, they are self-explanatory!
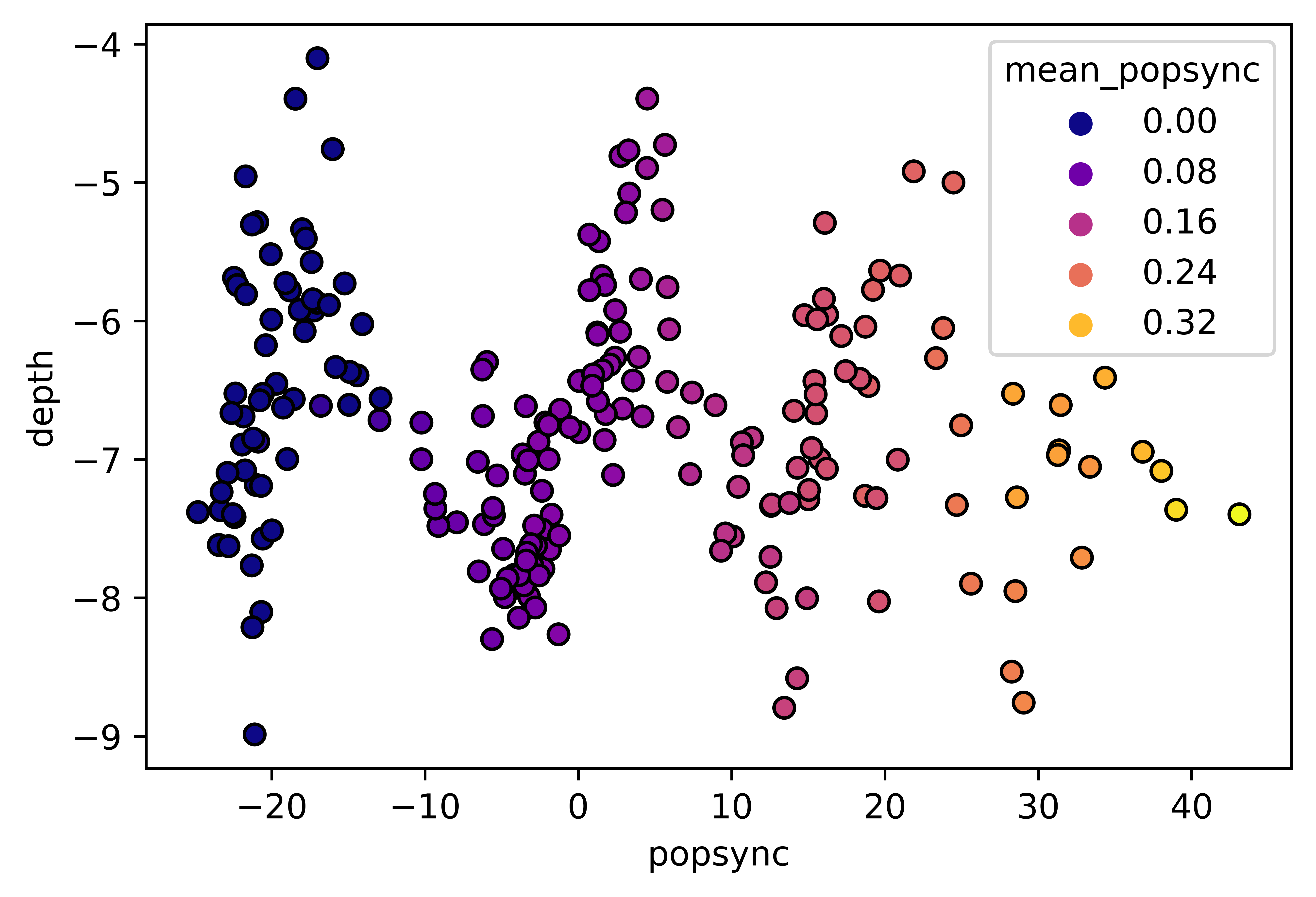
df1 = pd.load("my_dataframe.csv")
# Seaborn figure (seaborn is simply a wrapper for matplotlib):
fig = plt.figure()
sns.scatterplot(data=df1,
x='popsync', y='depth', hue='mean_popsync',
palette='plasma', alpha=1, linewidth=1, edgecolor='black')# Same figure, tweaked with mplp():
fig = plt.figure()
sns.scatterplot(data=df1,
x='popsync', y='depth', hue='mean_popsync',
palette='plasma', alpha=1, linewidth=1, edgecolor='black')
mplp(figsize=(3,3), title="My title", ylim=[-10,-2], xlim=[-40,60],
xlabel = "My x label (rotated ticks)", ylabel="My y label",
xtickrot=45,
hide_legend=True, colorbar=True,
vmin=df['mean_popsync'].min(), vmax=df['mean_popsync'].max(),
cbar_w=0.03, cbar_h=0.4, clabel="My colorbar label\n(no more ugly legend!)", cmap="plasma",
clabel_s=16, cticks_s=14, ticklab_s=16,
saveFig=saveFig, saveDir=saveDir, figname = f"popsync_{pair}")Using a conda/virtualenv environment is very much advised, as pre-existing packages on a python installation might be incompatible with npyx and break your installation (typically leading to python -c 'import npyx' failing). Instructions here: manage conda environments
Npyx supports Python >=3.7 <=3.10.
- as a user
- from pip (normally up to date)
conda create -n my_env python=3.10 conda activate my_env pip install npyx # either 11.0 or either cuda toolkit version you have already installed # find out with nvcc --version conda install -c conda-forge cupy cudatoolkit=11.0 # optional, for heavy preprocessing functions - see 'Dealing with cupy' section below python -c 'import npyx' # should not return any error # If it does, (re)install any missing (conflictual) dependencies with pip (hopefully none!) # and make sure that you are installing in a fresh environment.
- from the remote repository (always up to date)
conda create -n my_env python=3.10 conda activate my_env pip install git+https://github.com/m-beau/NeuroPyxels@master # either 11.0 or either cuda toolkit version you have already installed # find out with nvcc --version conda install -c conda-forge cupy cudatoolkit=11.0 # optional, for heavy preprocessing functions - see 'Dealing with cupy' section below python -c 'import npyx' # should not return any error # If it does, (re)install any missing (conflictual) dependencies with pip (hopefully none!) # and make sure that you are installing in a fresh environment.
- as a superuser (recommended if plans to work on it/regularly pull upgrades)
Tip: in an ipython/jupyter session, use
%load_ext autoreloadthen%autoreloadto make your local edits active in your session without having to restart your kernel. Amazing for development.and pull every now and then:conda create -n my_env python=3.10 conda activate my_env cd path/to/save_dir # any directory where your code will be accessible by your editor and safe. NOT downloads folder. git clone https://github.com/m-beau/NeuroPyxels cd NeuroPyxels pip install -e . # this will create an egg link to save_dir, which means that you do not need to reinstall the package each time you edit it (e.g. after pulling from github). # python setup.py develop deprecated (doesn't handle dependencies, unlike pip) # either 11.0 or either cuda toolkit version you have already installed # find out with nvcc --version conda install -c conda-forge cupy cudatoolkit=11.0 # optional, for heavy preprocessing functions - see 'Dealing with cupy' section below python -c 'import npyx' # should not return any error # If it does, (re)install any missing (conflictual) dependencies with pip (hopefully none!) # and make sure that you are installing in a fresh environment.
cd path/to/save_dir/NeuroPyxels git pull # And that's it, thanks to the egg link no need to reinstall the package! # be careful though: this will break if you edited the package. If you wish to contribute, I advise you # to either post issues and wait for me to fix your problem, or to get in touch with me and potentially # create your own branch from where you will be able to gracefully merge your edits with the master branch # after revision.
To run the preprocessing functions (borrowed from pykilosort), you will need NVIDIA drivers and cuda-toolkit installed on your computer. This can be the hardest part of the installation. To test if your is working OK you should be able to run the following:
nvidia-smi # Should show how much your GPU is being used right now
nvcc # This is the CUDA compiler
Errors with the CUDA installation can sometimes be fixed by downgrading the version of cudatoolkit installed. To check the current version run the following:
conda activate my_env
conda list cudatoolkit
To install version 10.0 for example run the following
conda activate my_env
conda remove cupy, cudatoolkit
conda install -c conda-forge cupy cudatoolkit=10.0
You can use the built-in unit testing function 'test_npyx' to make sure that npyx core functions run smoothly, all at once.
from npyx.testing import test_npyx
# any spike sorted recording compatible with phy
# (e.g. kilosort output)
dp = 'datapath/to/myrecording'
test_npyx(dp)
# if any test fails, enter interactive mode debug mode as follow:
%pdb ON # must be in a notebook or regular ipython session
test_npyx(dp, raise_error=True)--- npyx version 2.3.4 unit testing initiated, on directory /media/maxime/AnalysisSSD/test_dataset_artefact...
--- Successfully ran 'read_metadata' from npyx.inout.
--- Successfully ran 'get_npix_sync' from npyx.inout.
--- Successfully ran 'get_units' from npyx.gl.
--- Successfully ran 'ids' from npyx.spk_t.
--- Successfully ran 'trn' from npyx.spk_t.
--- Successfully ran 'trn_filtered' from npyx.spk_t.
--- Successfully ran 'wvf' from npyx.spk_wvf.
--- Successfully ran 'wvf_dsmatch' from npyx.spk_wvf.
--- Successfully ran 'get_peak_chan' from npyx.spk_wvf.
--- Successfully ran 'templates' from npyx.spk_wvf.
--- Successfully ran 'ccg' from npyx.corr.
--- Successfully ran 'plot_wvf' from npyx.plot.
--- Successfully ran 'plot_ccg' from npyx.plot.
--- Successfully ran 'plot_raw' from npyx.plot.
(bunch of plots...)
-
cannot import numba.core hence cannot import npyx
Older versions of numba did not feature the .core submodule. You probably run a too old version of numba, vestigial from an old installation - make sure that you install npyx in a fresh conda environment si that happens to you, and eventually that numba is not installed in your root:pip uninstall numba conda activate my_env pip uninstall numba pip install numba
- core dumped when importing
This seems to be an issue related to PyQt5 required by opencv (opencv-python). Solution:
# activate npyx environment first
pip uninstall opencv-python
pip install opencv-python
# pip install other missing dependencies
Full log:
In [1]: from npyx import *
In [2]: QObject::moveToThread: Current thread (0x5622e1ea6800) is not the object's thread (0x5622e30e86f0).
Cannot move to target thread (0x5622e1ea6800)
qt.qpa.plugin: Could not load the Qt platform plugin "xcb" in "/home/maxime/miniconda3/envs/npyx/lib/python3.7/site-packages/cv2/qt/plugins" even though it was found.
This application failed to start because no Qt platform plugin could be initialized. Reinstalling the application may fix this problem.
Available platform plugins are: xcb, eglfs, linuxfb, minimal, minimalegl, offscreen, vnc, wayland-egl, wayland, wayland-xcomposite-egl, wayland-xcomposite-glx, webgl.
Aborted (core dumped)
- I think I installed everything properly, but npyx is not found if I run 'python -c "import npyx" '!
Typically:
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
ModuleNotFoundError: No module named 'npyx'Make sure that the python installation that you are using is indeed the version of your new environment.
To do so, run "which python" on linux/mac or "where python" on windows: the output should be the path to the right environment e.g. "/home/.../anaconda/envs/npyx/bin/python". If it isn't, try to deactivate/reactivate your conda environment, or make sure you do not have conflicting python installations on your machine.
If you enjoy this package and use it for your research, you can:
- cite this github repo using its DOI: Beau, M., Lajko, A., Martínez, G., Häusser, M., & Kostadinov, D. (2021). NeuroPyxels: loading, processing and plotting Neuropixels data in python. Github, https://doi.org/10.5281/zenodo.5509776
- star this repo using the top-right star button.
Cheers!
Useful link to create a python package from a git repository
# ONLY ON DEDICATED BRANCH
cd path/to/save_dir/NeuroPyxels
git checkout DEDICATED_BRANCH_NAME # ++++++ IMPORTANT
git add.
git commit -m "commit details - try to be specific"
git push origin DEDICATED_BRANCH_NAME # ++++++ IMPORTANT
# Then pull request to master branch using the online github green button! Do not forget this last step, to allow the others repo to sync.First change the version in ./setup.py in a text editor
setup(name='npyx',
version='1.0',... # change to 1.1, 1.1.1...Then delete the old distribution files before re-generating them for the new version using twine:
rm -r ./dist
rm -r ./build
rm -r ./npyx.egg-info
python setup.py sdist bdist_wheel # this will generate version 1.1 wheel without overwriting version 1.0 wheel in ./distBefore pushing them to PyPI (older versions are saved online!)
twine upload dist/*
Uploading distributions to https://upload.pypi.org/legacy/
Enter your username: your-username
Enter your password: ****
Uploading npyx-1.1-py3-none-any.whl
100%|████████████████████████████████████████████████████████| 156k/156k [00:01<00:00, 96.8kB/s]
Uploading npyx-1.1.tar.gz
100%|█████████████████████████████████████████████████████████| 150k/150k [00:01<00:00, 142kB/s]