About・Abstract・Notes・Historical timeline・Current examples・Reusing data・ Future?・Epilog・See also

Public health emergencies require profound and swift action at scale with limited resources, often on the basis of incomplete information and frequently under rapidly evolving circumstances. While emergency-triggered sharing goes back millennia, the sharing of data and associated metadata is a relatively new flavour under this broader theme, but one that has been receiving steadily growing attention over the last few years, especially in the context of the Ebola or Zika outbreaks. By now, we have reached a point where data sharing must be considered a key component of addressing present, future and even past public health emergencies.
In response, researchers, research institutions, journals, funders and others have taken steps towards increasing the sharing of data around ongoing public health emergencies and in preparation for future ones. These measures range from the adoption of open lab notebooks to modifications of policies and funding lines, and they include conversations around infrastructure and cultural change.
In this contribution, I will provide an overview of different ways in which the sharing of data has played a role in public health emergencies, highlighting steps that have already been taken over the last decade as well as challenges still lying ahead.
While focusing on disease outbreaks, I will draw on examples from other public health emergencies as well (e.g. earthquakes or tropical storms) and discuss their applicability in the context of infections. The examples will span the entire data life cycle of public health emergencies, from preventive measures and routine public health surveillance data to the tracking of pathogens, investigating pathogen transmission and other host-pathogen interactions, as well as diagnostics, vaccination, epidemiological modelling, data ethics and other related topics, concluding with considerations around the potential impact of preserving and sharing data, or failing at that.
An Etherpad has been set up to allow for collaborative note taking and to facilitate discussion. It can be found at https://etherpad.wikimedia.org/p/Env-H-580-on-2019-01-31.
1854: Cholera in London・2002: SARS・2004: Indian Ocean earthquake・2005: H5N1・2005: Hurricane Katrina・2009: H1N1・2010: Haiti earthquake・2011: Tōhoku earthquake・2011: Escherichia coli O104:H4・2013: Typhoon Haiyan・2015: Ebola・2016: Zika
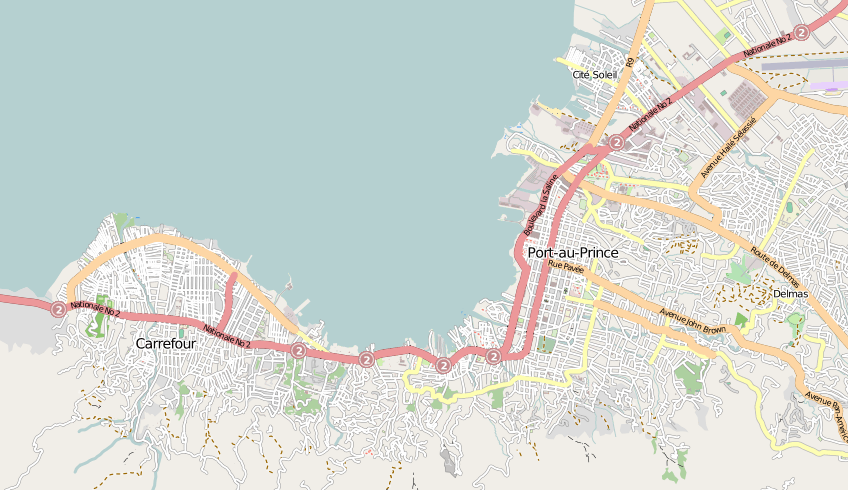
John Snow's original map of the 1854 Broad Street cholera outbreak in London. Cholera cases are highlighted in black, as are water pumps (data available here). The pump on Broad Street was identified as the one through which the contaiminated water was distributed. Removing its handle then essentially stopped the outbreak, and when the next Cholera outbreak hit London in 1866, sanitary measures had been improved.

- Anatomy of the Epidemiological Literature on the 2003 SARS Outbreaks in Hong Kong and Toronto: A Time-Stratified Review
-
Only 22% of the studies were submitted, 8% accepted, and 7% published during the epidemic.
-
Just as theoretical modelers have shifted to real-time approaches, for example, to estimate the basic/effective reproduction number of an epidemic [29], “field epidemiologists” should benefit from real-time tools for protocol writing and questionnaire design, and have them readily available on the Web so as to be prepared for the next emerging disease outbreak.
- editorial commentary
-
Based on their findings, it would be hard to conclude that journal publication was a successful mechanism for rapidly sharing information.
-
Whatever the answers may be, it seems clear that before the next public health emergency strikes, the scientific publishing establishment needs to ask itself how it can respond in the way the world needs.
-
-

- paper on mass fatality management
- from the editor's summary:
-
None of the countries had sufficient refrigerated storage available to store bodies until they could be identified.
-
Methods and efficiency of identification varied between and within countries. One hospital in Sri Lanka excelled by systematically photographing all bodies brought in and recording sex, height, and personal effects: 87% of the bodies brought here were identified. But in most areas rates of identification were much lower. It seemed that simple methods of identification were the most useful: photographs taken quickly before the bodies started to decompose, dental records, and personal effects found on the bodies. DNA analysis was only useful for a small number of bodies.
-
- from the editor's summary:

- Global Initiative on Sharing Avian Influenza Data (GISAID)
- paper
-
to understand better the spread and evolution of the virus, and the determinants of its transmissibility and pathogenicity in humans [..] demands that scientists with different fields of expertise have full access to comprehensive genetic-sequence, clinical and epidemiological data from both animal and human virus isolates.
-
The current level of collection and sharing of data is inadequate, however, given the magnitude of the threat.
-
We propose to expand and complement existing efforts with the creation of a global consortium — the Global Initiative on Sharing Avian Influenza Data (GISAID; http://gisaid.org) — that would foster international sharing of avian influenza isolates and data.
-
- paper


- PLOS Currents: Influenza was started: "The key goal of PLoS: Currents is to accelerate scientific discovery by allowing researchers to share their latest findings and ideas immediately with the world's scientific and medical communities."
- The ethics of sharing preliminary research findings during public health emergencies: a case study from the 2009 influenza pandemic
-
Cholera Epidemics of the Past Offer New Insights Into an Old Enemy
-
Mathematical modeling of the spread and health impact of cholera is a key effort to guide policy makers and intervention planners about the projected impact of interventions, such as vaccinations, in contemporary outbreaks [5–7].
-
Despite this public health importance, key aspects of cholera disease dynamics [..] contain a large amount of uncertainty or remain unresolved [8]. The high-quality epidemiological data needed to address these uncertainties are often lacking, especially in outbreak situations [6].
-
To fill this data void, we investigated data from an underutilized source: 19th-century cholera epidemics in Europe. Denmark provides an excellent source as its population was not exposed to cholera, likely due to a quarantine at the Danish coast [9]. Finally, in 1853, a year after the quarantine was lifted, a single and catastrophic outbreak hit the nation, including Copenhagen.
-
Time series of daily cholera morbidity and mortality counts by age and sex were obtained from datasets compiled by contemporary physicians in 3 towns and cities in Denmark: Copenhagen and Aalborg in 1853 and Korsør in 1857.
-
To provide epidemiological context to the outbreaks, we acquired cause-specific mortality data for the surrounding years for Copenhagen. We also obtained age-specific population data for Copenhagen, Aalborg, and Korsør.
-
-
Also in 2010: Sharing health data: good intentions are not enough
-
As they prepare for careers in science, today’s students doubtless hear the same clichés as we did a generation ago: science advances collaboratively; we reproduce and extend the work of others; we stand on the shoulders of giants. In some fields, such as genomics, these axioms are becoming true. In epidemiology and public health, however, data sharing and collaboration remain more aspirational than real.
-
Students embark on a career in health research in the spirit of sharing; they want to help improve the well-being of others. For all the talk of collaboration, they will enter a world in which another axiom dominates: “publish or perish”. That system puts the interests of public health researchers in direct conflict with the interests of public health.
-
Genomics has taught us that sharing data with other scientists is a way to add value without costing a lot. It allows the same data to be used to answer new questions that may be relevant far beyond the original study. And it allows for meta-analyses that are free from the distortions introduced when only summary results are available.3,4 We could get far more out of public health research if we followed a similar path, if we squeezed more scientific and policy insights out of data that have already been collected.
-
Sailors keeping log books on whaling boats in the 1600s could not have predicted that, centuries later, the data would be an important source of information for climate change scientists.25
-
Here we propose several goals to which funders and researchers can jointly aspire and towards which progress can be measured:
- (i) all data of potential public health importance funded by taxpayers or foundations will be appropriately documented and archived in formats accessible to the wider scientific community;
- (ii) all data provided by governments to databases developed by publicly-funded organizations will be freely available to any user, at the level of detail at which it was provided;
- (iii) the publication of a research article in a biomedical journal will be accompanied by the publication of the data set upon which the analysis is based;
- (iv) funders and employers of researchers will consider publication of well managed data sets as an important indicator of success in research, and will reward researchers professionally for sharing data; and
- (v) all planned research will budget and be funded to manage data professionally to a quality adequate for archiving and sharing.
-
Deaths and missing persons by prefecture from 2011 Tohoku Earthquake (by mti,InoueKeisuke,CES1596, public domain)
- OpenStreetMap coordination page
- Lots of people missing
- website sinsai.info set up by OpenStreetMap Japan using Ushahidi
- early version
- rough translation
- explanation
-
The dedicated site sinsai.info/ushahidi was established within two hours of the earthquake, to help search for survivors and provide vital information of safe spots and danger zones.
-
The site established by Japanese volunteers working with the Fletcher School at Tufts University, was already under construction in anticipation of an earthquake hitting Japan. The site enables anyone with a mobile phone or smartphone to post details of any survivors in difficult to reach areas and of any unsafe areas which is then relayed to rescue operations. In turn the site also posts easily accessible information on the nearest emergency services stations as well as locations of clean water supplies and food stores.
- early version
- website sinsai.info set up by OpenStreetMap Japan using Ushahidi
- saveMLAK
- Geigermap

- Open StreetMap coordination page
- OpenStreetMap-based poster maps printed in response to the typhoon were widely used by on-the-ground responders


-
Epidemiology
-
Data sharing
- Ebola teaches tough lessons about rapid research
- Data sharing: Make outbreak research open access
- Special issue of Journal of Empirical Research on Human Research Ethics: Ethics and sharing individual-level health research data from low and middle income settings (13 papers, 2015)
- Ebola outbreak data scraped from government PDF
- WHO Report of the Ebola Interim Assessment Panel - July 2015
-
Complex interactions between the Zika virus, humans and intermediates
-
- WHO Director-General declares Public Health Emergency of International Concern
-
I convened an Emergency Committee, under the International Health Regulations, to gather advice on the severity of the health threat associated with the continuing spread of Zika virus disease in Latin America and the Caribbean. The Committee met today by teleconference.
-
The experts agreed that a causal relationship between Zika infection during pregnancy and microcephaly is strongly suspected, though not yet scientifically proven. All agreed on the urgent need to coordinate international efforts to investigate and understand this relationship better.
-
The experts also considered patterns of recent spread and the broad geographical distribution of mosquito species that can transmit the virus.
-
I am now declaring that the recent cluster of microcephaly cases and other neurological disorders reported in Brazil, following a similar cluster in French Polynesia in 2014, constitutes a Public Health Emergency of International Concern.
-
The Committee found no public health justification for restrictions on travel or trade to prevent the spread of Zika virus.
-
- Fetal ultrasound
- MRI findings of microcephaly
- A Possible Link Between Pyriproxyfen and Microcephaly
- WHO Director-General declares Public Health Emergency of International Concern

Pool on an abandoned property (Idamantium, CC BY-SA 3.0)
- Vacant properties have been suggested to be breeding grounds for disease vectors, e.g. for Aedes aegypti
- Do vacant properties explain Miami's Zika outbreak? (follow-up post)
- uses "data collected by the U.S. Postal Service" to assess vacant properties.
- Do vacant properties explain Miami's Zika outbreak? (follow-up post)
- Such data correlates with poverty and housing prices, which could thus be another source information.

- Lots of discussion around data sharing
- Statement on data sharing in public health emergencies
- Policy Statement on Data Sharing by the World Health Organization in the Context of Public Health Emergencies
- Public Health Surveillance: A Call to Share Data
- Science, get over yourself: Zika data-sharing should be the norm, not the exception
- "Several participants noted that it is critical to study ZIKV in humans in the countries most affected and highlighted the importance of establishing a coordinated and well-resourced research approach to ZIKV, which would include the efficient sharing of biospecimens across international borders, availability of rapid funding announcements, better communication among scientists about the types of research being conducted, and the availability of datasets."
- Partnerships, Not Parachutists, for Zika Research:
-
But we believe the experience with recent outbreaks makes clear that if open sharing of data and specimens becomes the norm among scientists and epidemiologists around the world, we will be far more likely to succeed in improving international public health capacity and strengthening our collective health — and human — security.
-
To avoid having to make this argument again every time we face an outbreak with the potential for becoming a global crisis, we believe the global health community should develop and agree on a framework of principles for sharing data and biologic samples during any such public health emergency. It would be best if the researchers themselves developed such a framework, as the genomics community did in the Human Genome Project.
-
- Zika Open research Portal
- Nextstrain.org
- Open Zika
- Zika tracking
- Scholia on Zika virus
- other open datasets
- ContentMine: mining PubMed Central for data
- preprints
- from the Zibra blog (comparing Ebola versus Zika project): "there we were limited by upload speed and here throughput"
- Final Rule for FDAAA 801 and NIH Policy on Clinical Trial Reporting
- PCORI consultation on their data sharing policy

- press release by the Liberian Ministry of Health, announcing the finding on January 24, 2019:
“felt that this was an important finding to bring to the public irrespective of a scientific publication”
- only fragments of the viral sequence identified so far, so precise strain is as yet uncertain
- find was limited to a single sample out of > 11,000 that were analyzed
- sample was taken in 2016 from a Miniopterus inflatus bat in a cave in Liberia
- that species is common across multiple countries, but Ebola virus had never been identified in it in West Africa
- press release also highlights preventive measures and reiterates that there is no current outbreak in Liberia
- background: Liberia was the country most affected by the 2014 Ebola outbreak, which caught their health system by surprise, despite paywalled findings by foreign researchers "indicat[ing] that Liberia has to be included in the Ebola virus endemic zone" decades earlier
Q: What does Miniopterus inflatus feed on? A: Mostly insects.
Q: Where does Miniopterus inflatus live? A: In caves in various places across Sub-Saharan Africa.
Q: Are they migrating? A: Some closely related and equally insectivorous bat species are, e.g. the Natal long-fingered bat (Miniopterus natalensis).
Q: Am I allowed to embed their press release in this presentation? A: It's complicated.
 3D-printed Norovirus structure. NIH, CC BY 3D-printed Norovirus structure. NIH, CC BY |
 Serpentine wall in UVa's Academic Village. User:Uris, Public Domain Serpentine wall in UVa's Academic Village. User:Uris, Public Domain |
-
first brief news piece on January 30 (archived), simply informing about an email sent to UVa students, faculty and staff that alerted the community to the presence of the virus and provided some behavioral recommendations
-
follow-up later the same day (archived), highlighting preventive measures as outlined in the original email
-
Which institutions have affiliated researchers who published about Norovirus?
-
- how to find such samples taken in the past?
- similar case: 1970s and ‘Patient 0’ HIV-1 genomes illuminate early HIV/AIDS history in North America
- reused blood samples to sequence patient genomes to analyze the spread of HIV
- National Outbreak Recording System — collates data pertaining to various outbreaks and makes it searchable, which is helpful when attending to ongoing or future outbreaks, or even potential ones
- Zika data reuse
- An open RNA-Seq data analysis pipeline tutorial with an example of reprocessing data from a recent Zika virus study
- Jupyter notebook
- Docker image
- could in principle be run on a mobile phone
- Data-driven identification of potential Zika virus vectors
- How mobile data visualization helped reduce malaria cases by 93% — Zika could be next
- Mapping the global geographic potential of Zika virus spread
- Dynamic forecasting of Zika epidemics using Google Trends
- many Zika-themed hackathons
- Recent disasters — a Wikidata-based prototype to collect structured information about disasters worldwide
- Emergency Data Exchange Language (EDXL)
- Ontological issues
- Pathogen transmission ontology
- compare to Wikipedia article
- How does Friday 13th compare to other days in terms of probability of disaster?
- Make Data Sharing Routine to Prepare for Public Health Emergencies
- Big Data for Infectious Disease Surveillance and Modeling
- Global correlates of emerging zoonoses: Anthropogenic, environmental, and biodiversity risk factors
- Quantifying the global attention to public health threats through Wikipedia pageview data
- Data Management Plans for entire disease outbreaks and similar public health emergency scenarios
- Ten simple rules for machine-actionable data management plans
- take inspiration from infrastructure like the National Hurricane Center and expand internationally and beyond hurricanes
- Zika forest visit
- Satellite imagery to help disease surveillance
- Internet In a Box
- Livestream of data related to public health emergencies
- Your ideas here.


(Fire truck photo by Australian Department of Foreign Affairs and Trade, CC BY 2.0; data sharing illustration by Ainsley Seago, CC BY 4.0)
This file hosts a guest lecture given remotely on January 31, 2019, as part of the Environmental Health Seminar (Env H 580) at the Department of Environmental & Occupational Health Sciences at the University of Washington. It continues a series of talks on the subject that was started with a SciDataCon 2016 talk.
For information about the presenter, see here.
- Data sharing as a new component of addressing and preparing for disease outbreaks — a presentation given at SciDataCon 2018
- A guided tour through citation networks around public health emergencies
- Data sharing and public health emergencies — a presentation for Endangered Data Week 2018
- Data sharing in public health emergencies: A study of current policies, practices and infrastructure supporting the sharing of data to prevent and respond to epidemic and pandemic threats — a paper that I was notified of in response to a tweet about this talk.
- the same slides were also used for a presentation at the Institute of Geoinformatics of the University of Heidelberg on June 8, 2018
- Data sharing in public health emergencies — a presentation for SciDataCon 2016
- Data sharing in public health emergencies — a presentation for the International Meeting on Emerging Diseases and Surveillance (IMED 2016)
- More details on sharing in response to public health emergencies
- including the change log
- Problems
- Link rot
- Computational Biology Resources Lack Persistence and Usability
- Digital preservation of epidemic resources
- governmental documents
- "report from the National Science and Technology Council of the Executive Office of the President"
- search
- multiple examples highlighted throughout this talk (e.g. in the Tōhoku earthquake section)
- conclusion
- Data Centers affected by disaster
- low-quality metadata
- global applicability is not a given
- some potential data (e.g. on physiological adaptations, ecological interactions or chemical compounds) cannot be gathered because of biodiversity loss
- intentional removal by those who have control over the data
- ...
- Link rot
- Initiatives


