diff --git a/tutorials/misc/EEGLAB_and_iEEG_data.md b/tutorials/misc/EEGLAB_and_iEEG_data.md
index e309c9a..57263e3 100644
--- a/tutorials/misc/EEGLAB_and_iEEG_data.md
+++ b/tutorials/misc/EEGLAB_and_iEEG_data.md
@@ -5,29 +5,37 @@ long_title: EEGLAB and MEG data
parent: Reference Topics
grand_parent: Tutorials
---
-EEGLAB and iEEG/sEEG/ECoG data
+EEGLAB and iEEG, sEEG or ECoG data
====================
-EEGLAB supports reading most iEEG data formats (EDF, MEF3) through native code
-or plugins. The BIDS-matlab-tools EEGLAB plugin
-also support importing BIDS formatted MEG data. You may install plugins from the EEGLAB plugin manager (menu item File > Manage EEGLAB extensions).
+EEGLAB supports reading most iEEG data formats (EDF, MEF3, NWB) through native code
+or plugins. The BIDS-Matlab-tools EEGLAB plugin
+also supports importing BIDS-formatted MEG data. You may install plugins from the EEGLAB plugin manager (menu item File > Manage EEGLAB extensions).
-For example, after installing the MEF3 and BIDS-matlab-tools plugins, you may import the
+## Importing data
+
+For example, after installing the MEF3 and BIDS-Matlab-tools plugins, you may import the
[ds003708 BIDS dataset](https://nemar.org/dataexplorer/detail?dataset_id=ds003708&processed=0).
First, download the data. Second, use menu item File > BIDS Tools > Import BIDS folder to STUDY.
Leave all defaults and press OK (you may also select the column of interest for event types). Alternatively, use
menu item File > Import data > Using EEGLAB functions and plugins > Import MEF3 folder to import the mefd
folder located in the ds003708/sub-01/ses-ieeg01/ieeg/ folder of the BIDS dataset.
-When importing with BIDS, the advantage is that, along with the data, you will likely have access the iEEG electrode locations and relevant events. We show below the raw sEEG data for the unique subject in BIDS dataset ds003708.
+When importing with BIDS, the advantage is that, along with the data, you will likely have access to the iEEG electrode locations and relevant events. We show below the raw sEEG data for the unique subject in the BIDS dataset ds003708.
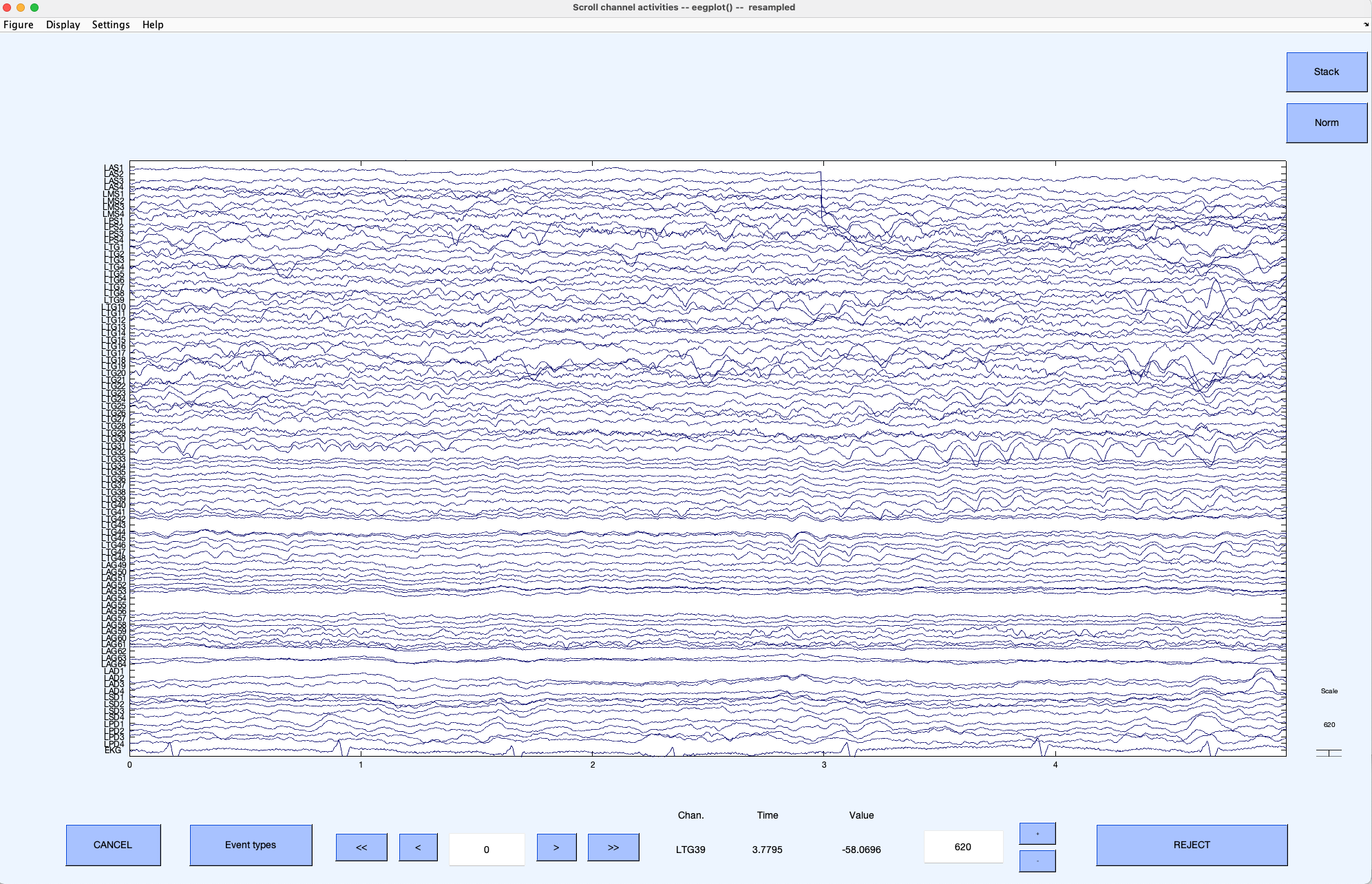
-Below the channel locations for the same dataset above are shown.
+The channel locations for the same dataset are shown below.

-Even if you are not planning to use EEGLAB to process iEEG data, importing your iEEG data into EEGLAB and resaving it into an EEGLAB dataset may be useful to process it in other software.
+Even if you are not planning to use EEGLAB to process iEEG data, importing your iEEG data into EEGLAB and resaving it into an EEGLAB dataset may be useful for processing it in other software.
+
+## Importing spike information
+
+EEGLAB allows importing spikes as events when using the NWB (Neurodata Without Border). Below, we show an example using the file [sub-01_ses-20140828T132700_ecephys+image.nwb](https://api.dandiarchive.org/api/assets/94ba06fc-c870-4698-9c31-f403ee733887/download/) of this [DandiSet](https://dandiarchive.org/dandiset/000576/). After downloading the file, and after installing the NWB-io EEGLAB plugins (menu item File > EEGLAB extensions then install the NWB-io plugin). After installing the plugin, import the file above in EEGLAB using menu item File > Import data > Using EEGLAB functions and plugins > From NWB file.
+
+Once the file has been imported, use menu item
Other relevant resources for processing iEEG data:
- [Fieldtrip sEEG tutorial](https://www.fieldtriptoolbox.org/tutorial/human_ecog/)