-
Notifications
You must be signed in to change notification settings - Fork 10
Architecture
Nacho edited this page Oct 23, 2015
·
8 revisions
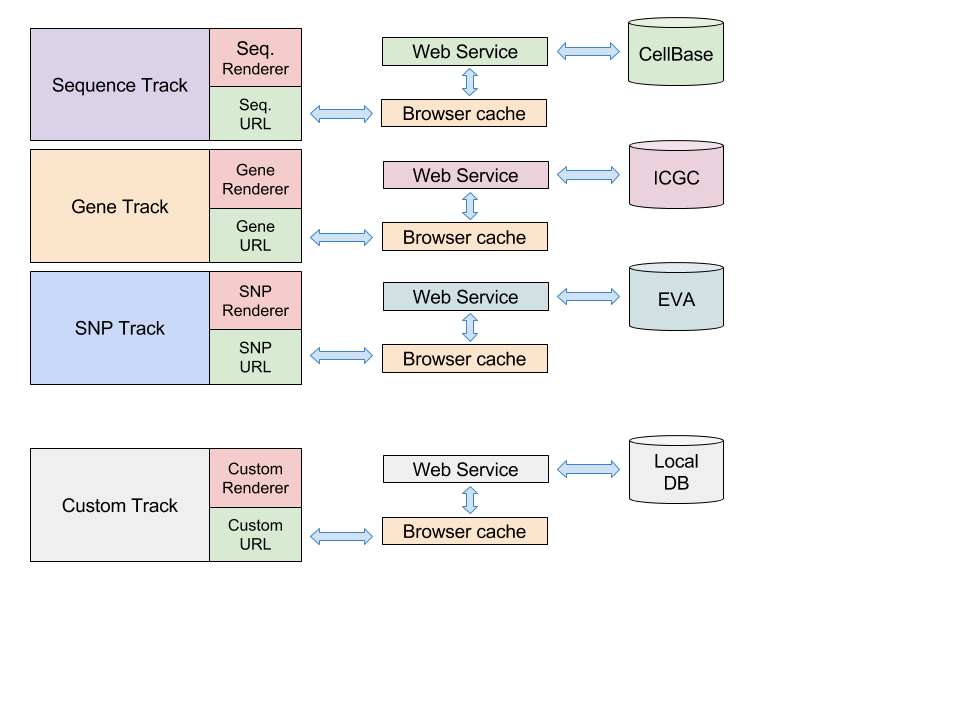
Genome Maps is the web-based genome browser and big data visualization tool of OpenCB platform, it sits on top of some other projects of OpenCB as can be seen in the next picture:

Genome Maps can render data from CellBase and OpenCGA projects:
- CellBase integrates public data repositories such as Ensembl into a NoSQL database and exports all this data through REST web services using well defined and specified data models. You can get some information about REST web services at https://github.com/opencb/cellbase/wiki/RESTful-web-services
- OpenCGA provides and authenticated environment to store and index BAM and VCF files -among others-, data is exported using REST web services as well to Genome Maps. OpenCGA Catalog is the responsible of authentication, authorization, sharing, ... You can get more information about web services at https://github.com/opencb/opencga/wiki/RESTful-Web-Services
Genome Maps has been designed to be highly modular and easy to integrate in other projects. Also, it is very simple to consume and render data from different sources such as ICGC or EMBL-EBI EVA.