-
Notifications
You must be signed in to change notification settings - Fork 10
Home
Genome Maps is an open source and high-performance HTML5 web-based genome browser. Genome Maps allows to browse several genomes and annotations by fetching data from CellBase high-performance REST web services. Custom data (BAMs, VCFs, ...) can also be loaded from remote servers and local disk by using OpenCGA server.
Genome Maps has been implemented to be very fast and designed in a modular way. All the web components and visualization technology developed for Genome Maps, and other related projects such as Cell Maps, are being developed in JSorolla JavaScript library. This project implements a JavaScript library for genomic and biological network data visualization and constitute the foundation of Genome Maps.
Genome Maps is written in JavaScript and makes an exhaustive use of the open standards HTML5 and CSS3; also, other open source libraries and technologies are used so Genome Maps runs in all modern web browsers without the installation of any web private technology. To take a quick look visit Genome Maps. You can find and cite the paper at Nuc. Acids Res. at http://nar.oxfordjournals.org/content/41/W1/W41.
Genome Maps allows a real-time navigation through big genomic regions:
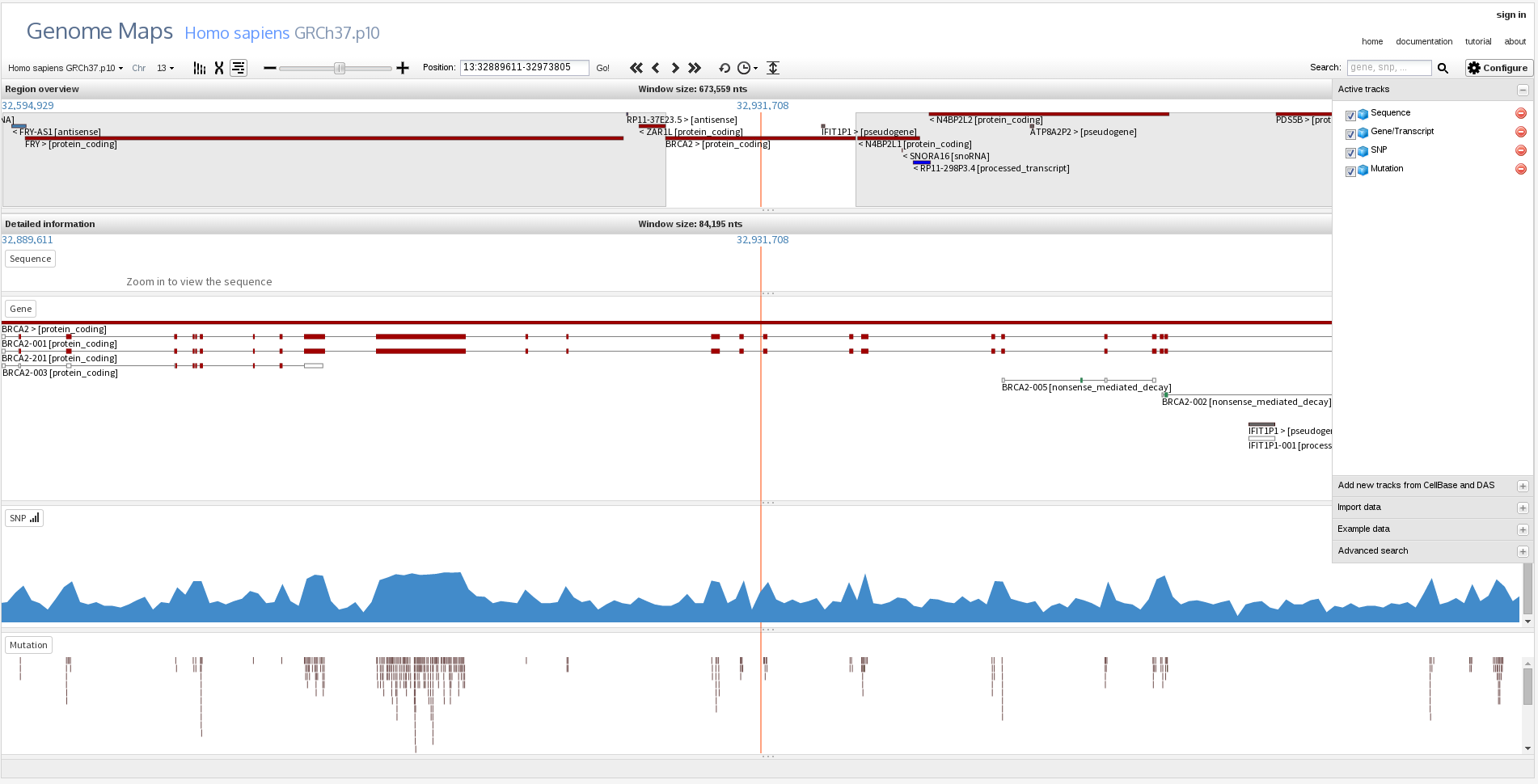
And also zoom in to feature or nucleotide level detail:
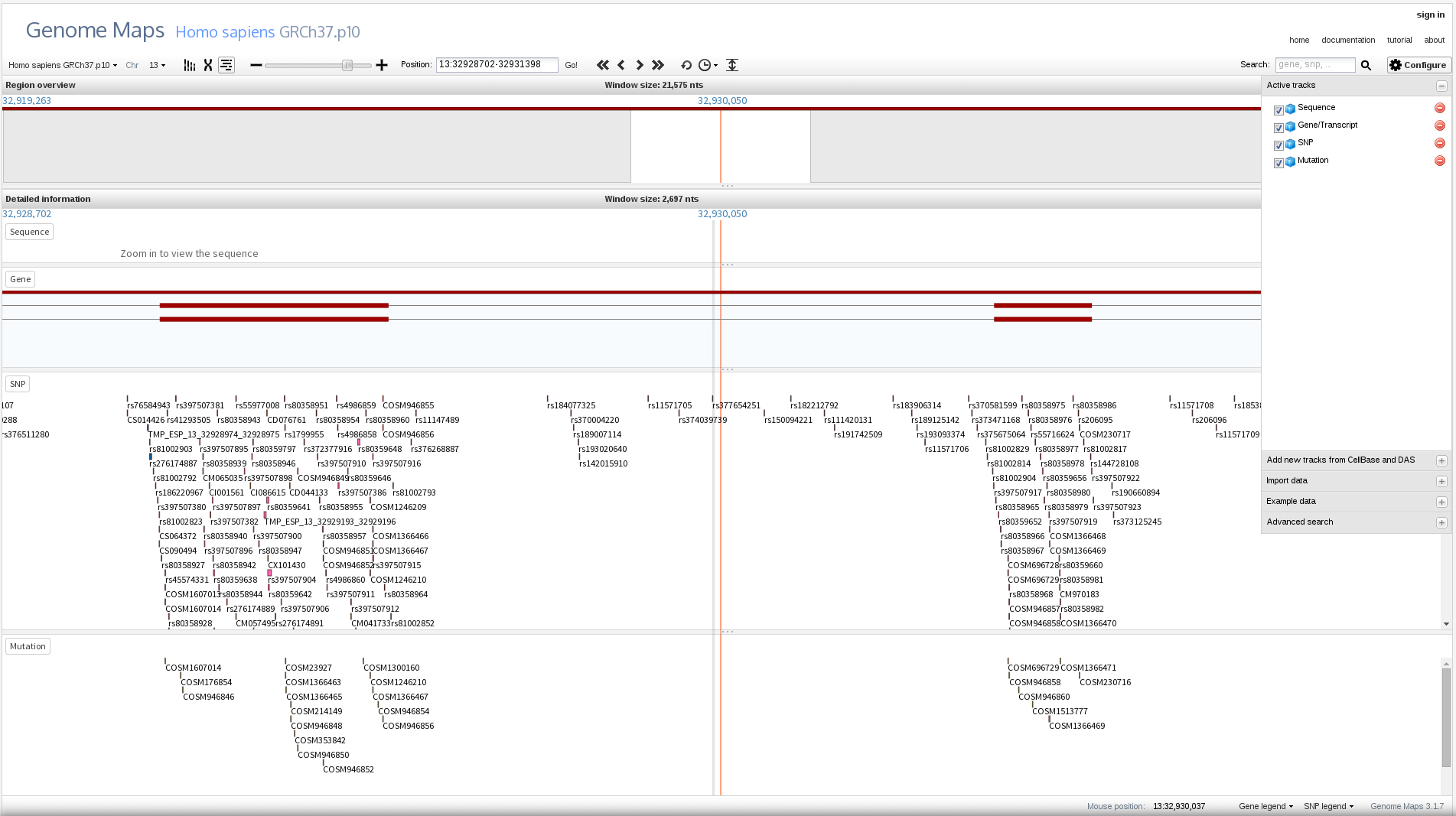
You can also easily choose among some built-in species and add more tacks with genome annotation from CellBase or even DAS servers, although these are usually to slow.
When OpenCGA is installed custom data files such as BAMs or VCFs can also be loaded from remote or local disk. Data files are first indexed to provide a quick navigation and filtering.
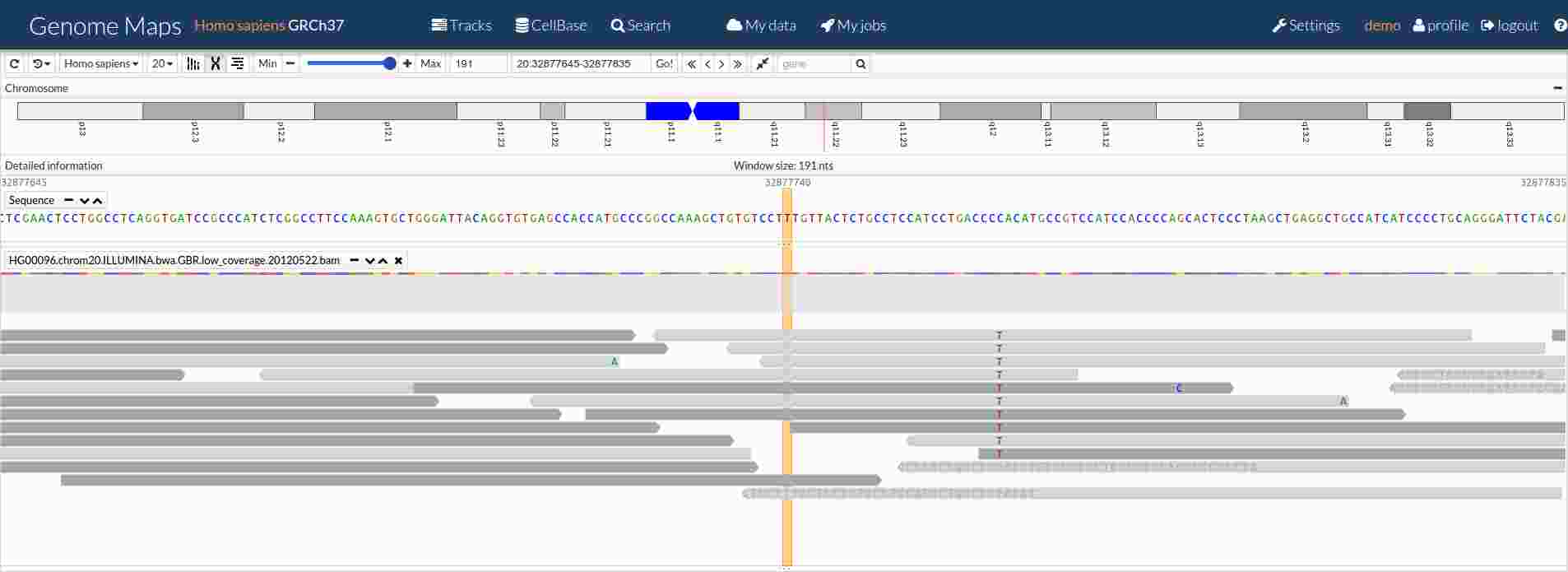 Genome Maps has been designed in a modular way and different components and APIs are provided. This makes very easy to reuse and customize some components. Source code is free and open source.
Genome Maps has been designed in a modular way and different components and APIs are provided. This makes very easy to reuse and customize some components. Source code is free and open source.
Genome Maps visualization technology is developed in an external project called JSorolla to easily be included in other projects. Today there are several projects using Genome Maps technology by importing JSorolla:
- The Genome Browser of ICGC project (https://dcc.icgc.org/search/g)
- Lens PatSeq Explorer (http://www.lens.org/lens/bio/patseqexplorer)
- Chaetomium thermophilum genome at EMBL (http://ct.bork.embl.de/ctbrowser)
- .. may more coming! If you are using please contact me!
Genome Maps project is led by Ignacio Medina ([email protected]) and begun in the Bioinformatics and Genomics Department led by Joaquin Dopazo ([email protected]) at CIPF (Valencia, Spain). Currently Ignacio Medina is working at University of Cambridge where he leads the Computational Biology Lab at HPC Service.
Two people deserve a special mention for their special contribution done: Franscisco Salavert which is currently the main developer and Alejandro de Maria who was the first developer.
Medina I., Salavert F., Sanchez R., de Maria A., Alonso R., Escobar P., Bleda M., Dopazo J. Genome Maps, a new generation genome browser. Nucleic Acids Res. 2013 Jul;41(Web Server issue):W41-6. doi: 10.1093/nar/gkt530
NAR - PubMed - [Google Scholar](https://scholar.google.co.uk/scholar?q=Genome+Maps%2C+a+new+generation+genome+browser&btnG=&hl=en&as_sdt=0