-
Notifications
You must be signed in to change notification settings - Fork 98
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Display cortical parcellation values: transparent medial wall and threshold both negative and positive values #290
Comments
I think this is because you are setting medial wall values to -1, when I think you want 0.
The
I'm not sure what you mean by "doesn't look good". |
|
My suggestion for your specific task is to use
I think the answer here is that the colormap has 256 entries and so if the distance between Also it's annoying that there's no way to use
I can't replicate this behavior on my system; the colormap maxes out at the maximum value in the data for all permutations of your examples. |
We could add a
Agreed we should try to rework the code if possible |
|
Ok I will use thresh at the lowest negative values above the value of medial wall. However, there is still a problem if I want to threshold the values symmetrically (both negative and positive values) to a particular min value. |
Not really; you can generalize the approach pretty easily: Where |


Hi,
I used on OSX
pysurfer=0.11.dev0withpython=3.6.10and tried to display on Desikan parcellation values, containing both negative and positives values, using the following code:And got the following result:
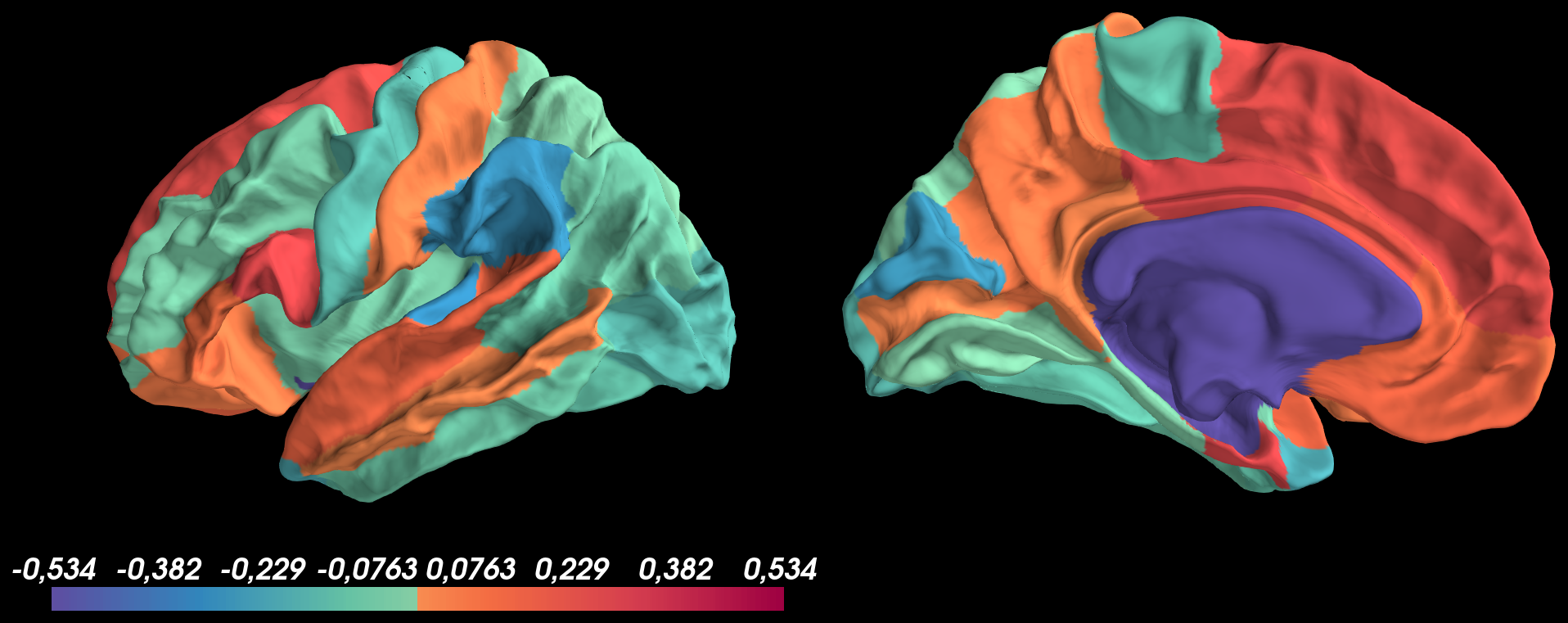
Based on this image, I don't understand why the medial wall isn't transparent?
Secondly, I tried to threshold both negative and positive values at
thresh_low = 0.1using pretty same code as before with thethreshoption ofadd_data:And got the following result:
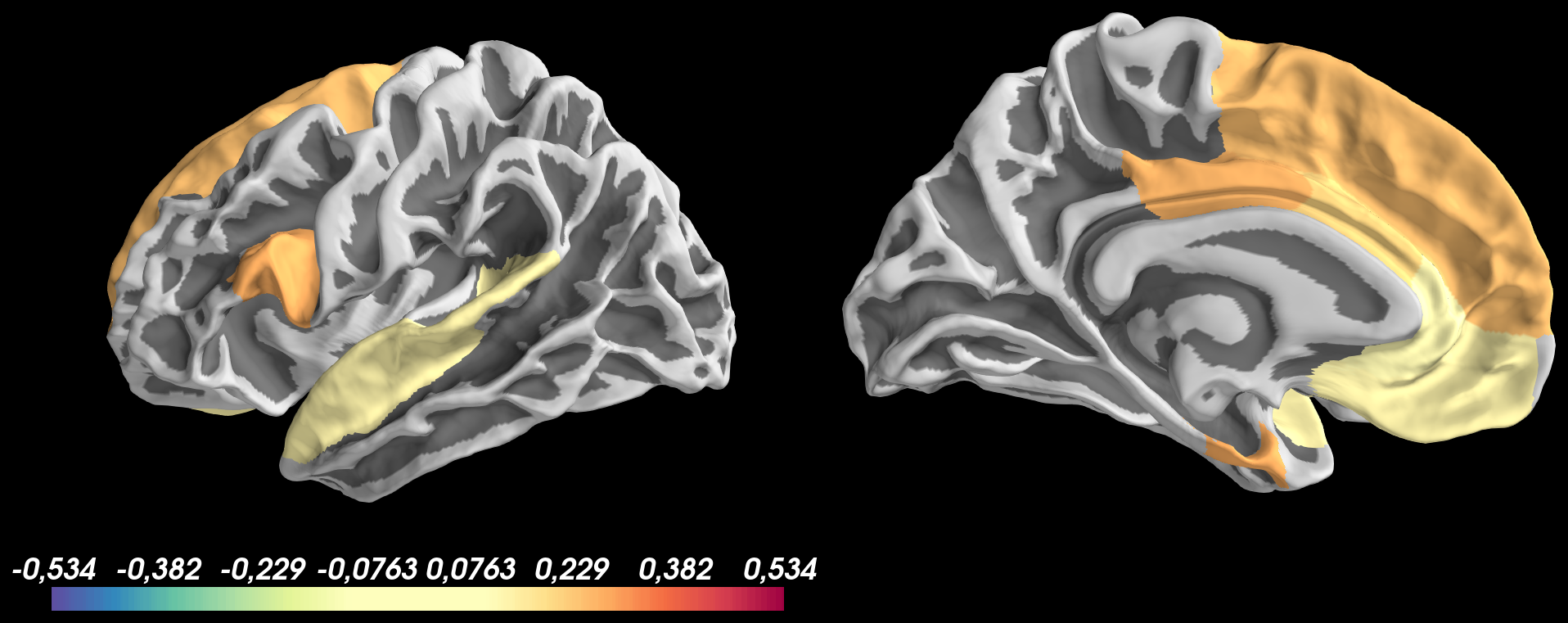
Based on this image, I don't understand why:
thresh_low = 0.1but negative values are not shown0.1 to 0.534for positive values and-0.1 to -0.534for negative valuesThe text was updated successfully, but these errors were encountered: