-
Notifications
You must be signed in to change notification settings - Fork 8
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
How to use peak picking in processed spectra
#6
Comments
|
This type of binning might produce tall but jagged peaks which would violate the signal to noise ratio threshold. Without seeing the actual values in the upsampled arrays, I couldn't say what's actually going on though. Alignment could be the issue. If you want to merge profile spectra, you can use import ms_peak_picker
spectra = [(mz_array, intensity_array) , (mz_array2, intensity_array2), ...]
mz_grid, intensity_average = ms_peak_picker.average_signal(spectra, dx=0.001)
intensity_average *= len(spectra) # scale the average by the number of spectra to convert to the sum of spectra
mz_grid, intensity_smoothed = ms_peak_picker.scan_filter.SavitskyGolayFilter()(mz_grid, intensity_average)
peak_list = ms_peak_picker.pick_peaks(mz_grid, intensity_smoothed)If you want average peak lists, you can use import ms_peak_picker
spectra = [peaks1, peaks2, ...]
mz_grid, intensity_average = ms_peak_picker.reprofile(spectra, dx=0.001)
intensity_average *= len(spectra) # scale the average by the number of spectra to convert to the sum of spectra
mz_grid, intensity_smoothed = ms_peak_picker.scan_filter.SavitskyGolayFilter()(mz_grid, intensity_average)
peak_list = ms_peak_picker.pick_peaks(mz_grid, intensity_smoothed) |
After with My goal is to compare the spectra, so I actually may not need average spectra. |
|
To be precise, you do not wish to have centroids, but a 2-D matrix where each row is a spectrum and each column is the intensity at a specific m/z shared across all spectra? import pickle
import numpy as np
from scipy.spatial import distance
import ms_peak_picker
%matplotlib inline
from matplotlib import pyplot as plt
from ms_peak_picker.plot import draw_raw, draw_peaklist
plt.rcParams['figure.figsize'] = 10, 6
# Load spectra
spectra = pickle.load(open("./rawmzspectra1.bin", 'rb'))
# Rebin with linear interpolation without merging or any other transformation.
binner = ms_peak_picker.scan_filter.LinearResampling(0.001)
rebinned = [binner(*s) for s in spectra]
# Prove that all m/z axes are identical
for i, ri in enumerate(rebinned):
if i < len(rebinned) - 1:
rj = rebinned[i + 1]
else:
rj = rebinned[0]
assert np.allclose(ri[0], rj[0])
# Construct the intensity matrix and store a copy of the shared m/z axis
mz_grid = rebinned[0][0]
intensity_grid = np.vstack([ri[1] for ri in rebinned])
# Example pairwise cosine similarity
print(distance.cosine(intensity_grid[0, :], intensity_grid[1, :]))
# Confirm that peak signal is broadly the same location but different magnitudes
ax = draw_raw(mz_grid, intensity_grid[0, :], alpha=0.5)
draw_raw(mz_grid, intensity_grid[1, :], alpha=0.5, ax=ax)
plt.savefig("spectrum_comparison.pdf")
# Compute pairwise distance matrix over all spectra
distmat = distance.squareform(
distance.pdist(intensity_grid, metric='cosine', )
)
# Plot pairwise distance matrix as a heat map
q = plt.pcolormesh(distmat)
cbar = plt.colorbar(q)
cbar.ax.set_ylabel("Cosine Similarity", rotation=270, size=12, labelpad=15)
_ = plt.title("Cosine Similarity Pairwise Matrix", size=16)
_ = plt.xticks(np.arange(0.5, 10.5), np.arange(0, 10))
_ = plt.yticks(np.arange(0.5, 10.5), np.arange(0, 10)) |
|
Yes, I want the data structure to be a 2D matrix as you've mentioned with a uniform m/z grid. Moreover, the equal spacing/step binning does not preserve the distribution of m/zs. The blue vertical lines are centroids. Is there a way to uniform/homogenize the spectra with |
|
The spacing argument of the You could then use import math
from collections import defaultdict
def sparse_peak_set_similarity(peak_set_a, peak_set_b, precision=0):
"""Computes the normalized dot product, also called cosine similarity between
two peak sets, a similarity metric ranging between 0 (dissimilar) to 1.0 (similar).
Parameters
----------
peak_set_a : Iterable of Peak-like
peak_set_b : Iterable of Peak-like
The two peak collections to compare. It is usually only useful to
compare the similarity of peaks of the same class, so the types
of the elements of `peak_set_a` and `peak_set_b` should match.
precision : int, optional
The precision of rounding to use when binning spectra. Defaults to 0
Returns
-------
float
The similarity between peak_set_a and peak_set_b. Between 0.0 and 1.0
"""
bin_a = defaultdict(float)
bin_b = defaultdict(float)
positions = set()
for peak in peak_set_a:
mz = round(peak.mz, precision)
bin_a[mz] += peak.intensity
positions.add(mz)
for peak in peak_set_b:
mz = round(peak.mz, precision)
bin_b[mz] += peak.intensity
positions.add(mz)
return bin_dot_product(positions, bin_a, bin_b)
def bin_dot_product(positions, bin_a, bin_b, normalize=True):
'''Compute a normalzied dot product between two aligned intensity maps
'''
z = 0
n_a = 0
n_b = 0
for mz in positions:
a = bin_a[mz]
b = bin_b[mz]
z += a * b
n_a += a ** 2
n_b += b ** 2
if not normalize:
return z
n_ab = math.sqrt(n_a) * math.sqrt(n_b)
if n_ab == 0.0:
return 0.0
else:
return z / n_abThis method doesn't even require you to homogenize the m/z axis. You select the level of granularity by raising or lowering the The KDE plot you showed is either using a bandwidth that is way too broad or you're trying to summarize big chunks of the m/z dimension together intentionally. I can't think of a reason you'd want to do that with high resolution spectra. I don't understand why the |
|
Hello Joshua,
Sorry for the confusion. I was actually not looking to compare separate spectrums. I could do that with The reasons to homogenize the spectra to a 2D array with same mzgrid are to:
In general the above generates 1603 and 1749 peaks each. That looks good as I can fetch individual fitted peaks and their properties( Is there a way to align the peaks or m/zs with the
One other question, in this context since we are taking one intensity value per m/z into consideration, how significant the width of peaks is?
Yes, you are right, the KDE plot comes from an optimized bandwidth. I got the concept from this paper that uses the KDE generated clusters for Peak Detection and Filtering Based on Spatial Distribution. I am relatively new in MSI data so have a bit of questions, sorry for that. |
|
Hello Joshua, The final matrix size is 10x4251 which I am pretty happy with.
this was done to avoid noisy peaks, as avg intensity of spectra was multiplied by
|
|
I may have missed something early on after seeing the spectra you shared were profile mode. Are you trying to do this with pre-centroided spectra? |
|
To confirm: is centroided mode where the spectra are like impulses at the center of the profile peak? When generating ion images/downstream analysis, shouldn't it be a single centroided value per m/z? Could you explain what mode should be chosen in this case and are there specific applications for that? |
|
Yes, a centroid spectrum have already been reduced to the discrete centers of a profile peak, and ideally no zero intensity points. Downstream analyses should use centroided spectra, but attempting to put two centroid spectra on the same axis to generate a shared intensity map involves binning those discrete centroids or converting them back into profiles on a shared m/z axis. When you give When the If you pass |
|
Thanks for the detailed explanation. I want to show you 3 methods: Obviously, visually it seems the prominence one seems to follow the spectrum(#8) with computationally lower number of peaks.
|
|
What are you doing to produce the data array you're plotting here? Using import numpy as np
import ms_peak_picker
import pickle
from matplotlib import pyplot as plt
plt.rcParams['figure.figsize'] = 12, 6
data = pickle.load(open("./rawmzspectra1.bin", 'rb'))
for i, (mz, intensity) in enumerate(data):
peaks = ms_peak_picker.pick_peaks(mz, intensity, signal_to_noise_threshold=7, peak_mode='profile')
ax = ms_peak_picker.plot.draw_raw(mz, intensity, color='red', label='Raw')
ms_peak_picker.plot.draw_peaklist(peaks, ax=ax, color='blue', label='Picked Peaks', lw=0.5)
ax.legend()
ax.set_title(f"Index {i}")
plt.savefig(f"{1}.png", bbox_inches='tight')
plt.close(ax.figure) |
|
I peak picked with the |



















I have a different number of m/zs per pixel/spectrum(processed). How should I use this tool to homogenize all the spectra?
Currently, I am binning and upsampling(summing all intensity in between bins) all the spectra using step size 0.01/0.02 which roughly ends up being
130,000m/zs per spectrum. Then I take the average of the spectra and perform peak picking on that to find peak indices...refmz= common mass m/z range with step 0.01.meanSpec= mean of spectra after upsampling.I am using the following parameters:
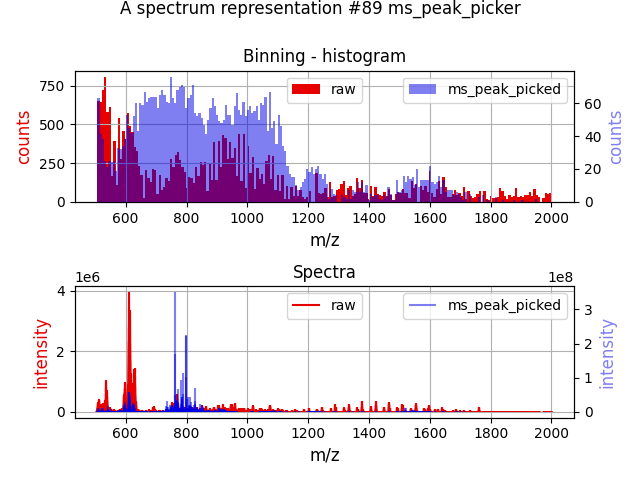
picking_method="quadratic", snr=10, intensity_threshold=5, fwhm_expansion=2But the resultant spectrum doesn't look anywhere near the raw spectrum if plotted.
Moreover, is there any alignment required additionally?
The text was updated successfully, but these errors were encountered: