Effect model of gene-environment interaction, involved in predictive analysis of GWAS and GS

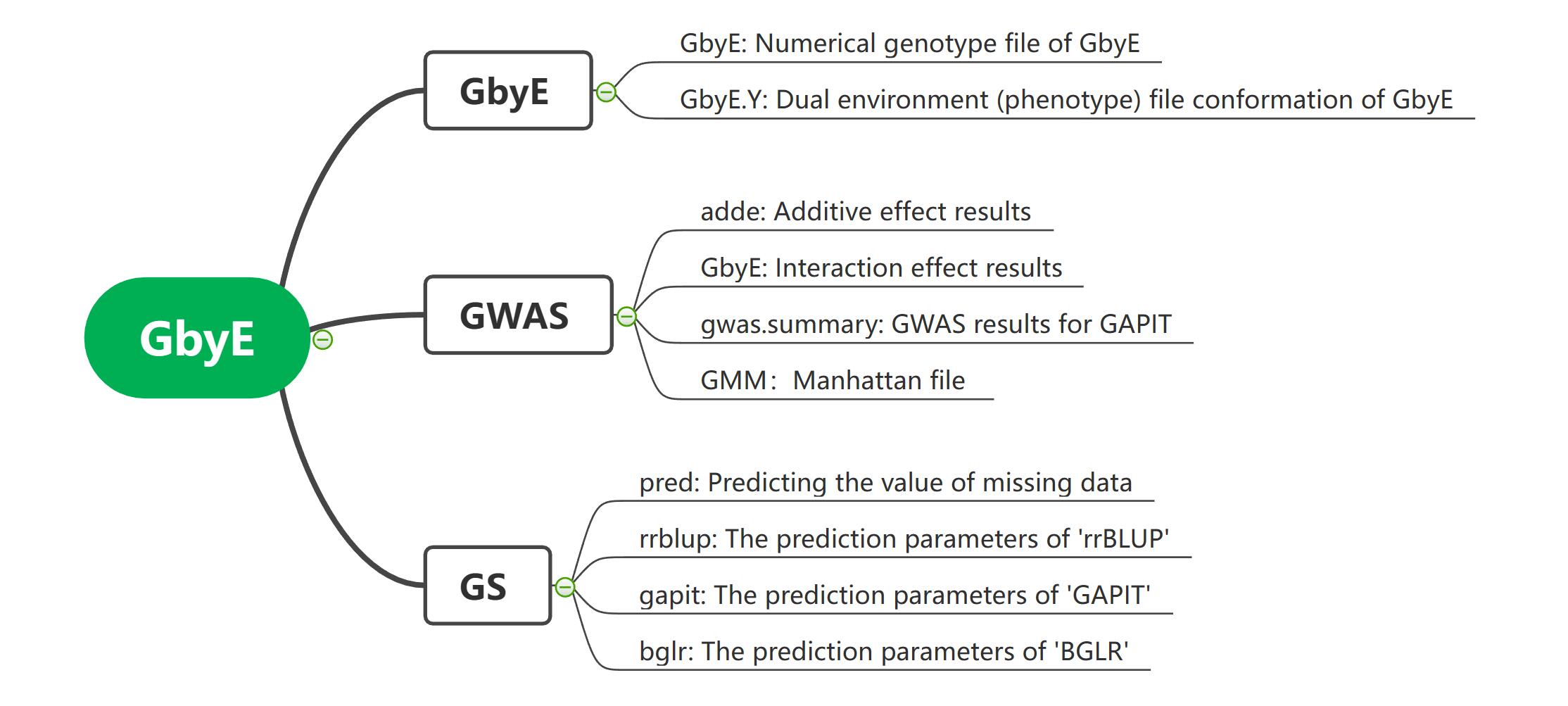
Gbye main package, coordinate other function packages for GWAS and GS operations, sometimes, We need to prepare documents, including
GD(Genotype file,type [0,1,2])GM(SNP marker map file with SNP names, Chrosome and Position)Y(Phenotype file,The phenotype file must contain two environments or two traits)GbyE.GD&GbyE.GM(If you have previously performed GbyE operations and saved a GbyE file, you can add the file directly to GbyE to reduce the time cost incurred by calculating again)gwas(defult=T, Whether to perform GWAS operation)PCA.total(Number of principal components as covariates at GWAS)CV(GbyE is based on GAPIT operations, and like GAPIT, it runs with covariates for operations)gwas.model(Select the GWAS analysis model you need, theoretically you can use all GAPIT models, such as: 'MLM', 'CMLM', 'BLINK', 'MLMM')cutOff(Allows setting a threshold of significance when drawing)gs(defult=T, Whether to perform GS operation)method(Set the operation method of GS, currently only 'BGLR', 'rrBLUP' and 'GAPIT' are supported)gs.model(GS model: rrblup-default-"ML", gapit "gblup" "mablup" "cblup" "sblup", bglr "RRB" "BA" "BC" "BB" "BL")nIter&burnIn(set Markov chain length of the R packages "BGLR")file.output(Whether the operation result file is output)plot(Whether curve drawing is carried out on the results can show the prediction accuracy of the results)file.type(Set the output image format, the default is PDF)dpi(Set the resolution of the output image, default=600)
The output results contain three panels, GbyE, GWAS and GS. GbyE mainly includes the output GbyE genotype file and phenotype file. GWAS mainly includes additive effect adde, reciprocal effect GXE, overall summary result of GWAS, GMM mapping parameter file. GS contains mainly, prediction results of all missing values pred. The prediction parameters of rrBLUP. The prediction parameters of GAPIT software, gapit. The prediction parameters of BGLR software, bglr.
Gbye main package, coordinate other function packages for GWAS and GS operations, sometimes, We need to prepare documents, including
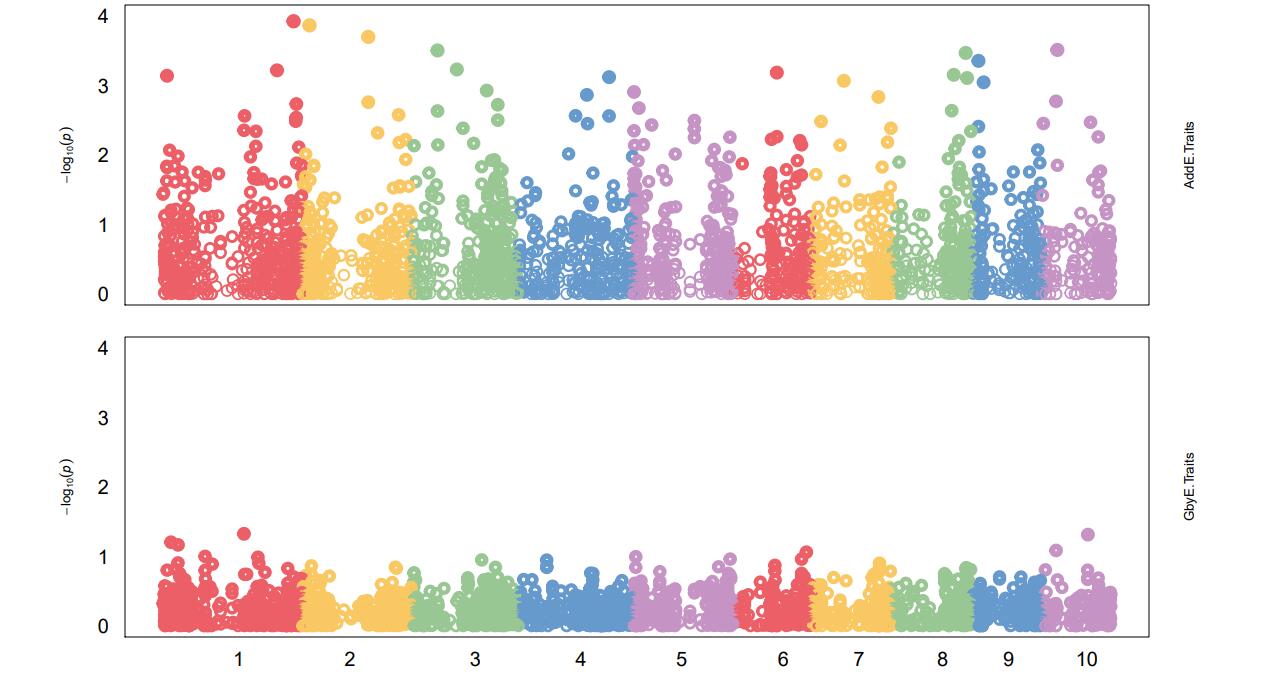
Manhattan Plots
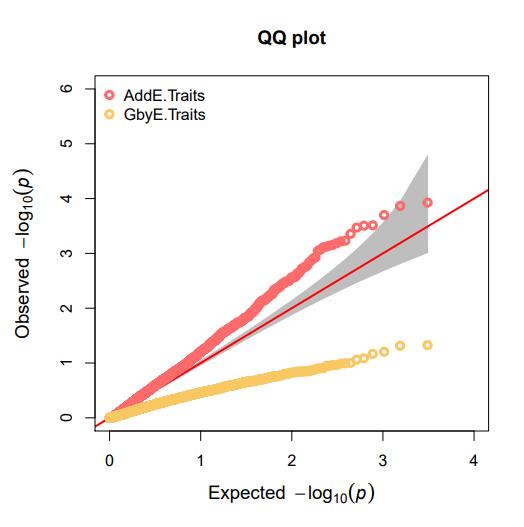
 Quantile-Quantile Plots
Quantile-Quantile Plots
#Loading support packages
source("https://raw.githubusercontent.com/liu-xinrui/GbyE/main/GbyE.R")
source("https://raw.githubusercontent.com/liu-xinrui/data/main/gapit_functions.txt")
source("https://raw.githubusercontent.com/liu-xinrui/data/main/GAPIT.library.R")
#Please import your own data below
GD=read.table("mdp_numeric.txt",head=T)
GM=read.table("mdp_SNP_information.txt",head=T)
Y=read.table("mdp_traits.txt",head=T)
#Remove phenotypic data without genotypes to maintain data consistency
Y=Y[match(GD[,1],Y[,1]),]
#Run GbyE
myGbyE=GbyE(GD=GD,
GM=GM,
Y=Y,
PCA.total=3,
gwas=T,
gs=T,
plot=T,
gwas.model="MLM",
method="gapit")
#GAPIT-" MLM" " CMLM" ,"BLINK" "FramCPU" "SUPER" etc
GbyE_GWAS=GbyE(GD=GD,
GM=GM,
Y=Y,
PCA.total=3,
gwas=T,
plot=T,
gwas.model=c("MLM","CMLM","BLINK","FramCPU","SUPER"))
#Support all rrblup built-in methods
GbyE_GS_rrBLUP=GbyE(GD=GD,
GM=GM,
Y=Y,
gs=T,
method="rrblup",
gs.model="ML")
#Support all GAPIT built-in methods
GbyE_GS_GAPIT=GbyE(GD=GD,
GM=GM,
Y=Y,
gs=T,
method="gapit",
gs.model=c("gBLUP","MABLUP","cBLUP","sBLUP"))
#Support all rrblup built-in methods
GbyE_GS_BGLR=GbyE(GD=GD,
GM=GM,
Y=Y,
gs=T,
method="bglr",
gs.model="RRB") #BL BA BB BC RRB etc
Here is a small function package related to gbye operation, which helps to run gbye programs more conveniently
This currently includes:
The following is used to support GbyE runs and to run tests and refine GbyE builds. unlike the main GbyE function package, GbyE.test.R adds simulation functions for testing GbyE only. the following are the parameters of this function, including
GD(Genotype file,type [0,1,2])GM(SNP marker map file with SNP names, Chrosome and Position)Y(Phenotype file,If the model phenotype is not used and the real phenotype prediction is directly used, please enter this file)ha2(Heritability of the simulate phenotype)cov_g(Genetic correlation of the simulate phenotyp)NQTN(Number of phenotype related SNPs simulated, defult=20)nrep(Number of iterations)nfold(CV Multiple cross validation)nIter&burnIn(set Markov chain length of the R packages "BGLR")gwas(defult=T, Whether to perform GWAS operation)gs(defult=T, Whether to perform GS operation)file.output(Whether the operation result file is output)plot(Whether curve drawing is carried out on the results can show the prediction accuracy of the results)
Function package for phenotypic simulation according to genotype file
Gbye file is generated from the original genotype file and SNP map file to show the effect of gene environment interaction
Calculate the FDR and Power values of the prediction results
In GWAS operation, compare the pvalue value of the same SNP and screen the most significant p value for subsequent GS prediction