Bender offers set of methods to handle TisTos-information in (relatively) easy way.
This functionality comes from BenderTools.TisTos module.
In short, it adds three relates method in the base class Algo:
| Method | Short description |
|---|---|
decisions |
collect the trigger decisions for the given particle |
trgDecs |
print the collected trigger statistics in readable way |
tisTos |
fill N-tuple with TisTos-information |
All of them are (relatively well) documented and one can easily inspect them:
import BenderTools.TisTos
from Bender.Main import Algo
help(Algo.decisions)Often information about the releavant trigger lines are spread in corridors
in a form of myths, general beliefs or, in the best case, references to ANA-notes
for some similar analysis. However it is very simple to collect
this infomation using Bender. The method decisions is our friend here.
the usage of this method require some preparatory work for the algorithm,
namely one needs to instrument initialize-method:
class TrgLines(Algo):
"""Collect infomation about the trigger lines relevant for certain decays/particles
"""
def initialize ( self ) :
sc = Algo.initialize ( self ) ## initialize the base class
if sc.isFailure() : return sc
#
## container to collect trigger information, e.g. list of fired lines
#
triggers = {}
triggers ['psi'] = {} ## slot to keep the information for J/psi
triggers ['K'] = {} ## slot to keep the information for kaons
triggers ['B'] = {} ## slot to keep the information for B-mesons
sc = self.tisTos_initialize ( triggers , lines = {} )
if sc.isFailure() : return sc
return SUCCESSHere in this example we want to collect TisTos-information
for J/psi, K and the whole B+-meson. Then in the main analyse-method one just needs to invoke
the method decisions for each particles in interest:
def analyse( self ) : ## IMPORTANT!
"""The main 'analysis' method """
...
for b in particles :
psi =b(1) ## the first daughter: J/psi
k =b(2) ## the second daughter: K
## collect trigger information for J/psi
self.decisions ( psi , self.triggers['psi'] )
## collect trigger information for kaons
self.decisions ( k , self.triggers['K'] )
## collect trigger information for B-mesons
self.decisions ( b , self.triggers['B'] )
...
return SUCCESS Thats all. Then when jobs runs it dumps to the log-file the running trigger statistics,
and the statistics is dumped into the file TrgLines_tistos.txt (<ALGNAME>_tistos.txt in general).
The summary table looks like:
******************************************************************************************
Triggers for psi
******************************************************************************************
Hlt1_TIS psi #lines: 7 #events 321
( 0.62 +- 0.44 ) Hlt1DiMuonHighMassDecision
( 0.62 +- 0.44 ) Hlt1DiMuonLowMassDecision
( 19.00 +- 2.19 ) Hlt1TrackAllL0Decision
( 3.74 +- 1.06 ) Hlt1TrackAllL0TightDecision
( 4.36 +- 1.14 ) Hlt1TrackMuonDecision
( 0.93 +- 0.54 ) Hlt1TrackPhotonDecision
( 0.31 +- 0.31 ) Hlt1VertexDisplVertexDecision
(100.00 +- 0.31 ) TOTAL
Hlt1_TOS psi #lines: 9 #events 321
( 72.90 +- 2.48 ) Hlt1DiMuonHighMassDecision
( 59.81 +- 2.74 ) Hlt1DiMuonLowMassDecision
( 0.31 +- 0.31 ) Hlt1DiProtonDecision
( 9.03 +- 1.60 ) Hlt1SingleMuonHighPTDecision
( 0.31 +- 0.31 ) Hlt1SingleMuonNoIPDecision
( 44.24 +- 2.77 ) Hlt1TrackAllL0Decision
( 10.90 +- 1.74 ) Hlt1TrackAllL0TightDecision
( 74.77 +- 2.42 ) Hlt1TrackMuonDecision
( 0.31 +- 0.31 ) Hlt1TrackPhotonDecision
(100.00 +- 0.31 ) TOTAL
Hlt1_TPS psi #lines: 6 #events 321
( 13.71 +- 1.92 ) Hlt1DiMuonHighMassDecision
( 10.90 +- 1.74 ) Hlt1DiMuonLowMassDecision
( 0.31 +- 0.31 ) Hlt1DiProtonDecision
( 3.12 +- 0.97 ) Hlt1TrackAllL0Decision
( 0.93 +- 0.54 ) Hlt1TrackAllL0TightDecision
( 4.67 +- 1.18 ) Hlt1TrackMuonDecision
(100.00 +- 0.31 ) TOTAL
Only a short fragment is shown here, one gets similar fragments for all declared
particles (psi, K and B) and for all trigger levels (L0, Hlt1 and Hlt2).
The full table is accessible here
The content of the summaty table is rather intuitive: it summarizes the fire frequences for
varios trugegr lines for three regimes TIS, TOS and TPS.
Inspecting such table, one immediately concludes that the most relevan Hlt1-TOS-line
is Hlt1DiMuonHighMassDecision. Other Hlt1-TOS-lines are less relevant here.
But please note that here only very small statistics is used (321 event),
and with larger statistics sthe conclusions could be corrected.
E.g. due to small statistics here, for Hlt1-TIS-lines the choice is not evident:
one clearly see that Hlt1TrackAllL0Decision line is important, but for
importance of other lines one csn judge only after the
significant increase of the statistics.
{% discussion "What is <ALGNAME>_tistos.db file?" %}
In practice to make a decision, large statistics is required (for real data and/or for simulated samples).
And here these files are very useful.
The trigger statistics is saved not only in <ALGNAME>_tistos.txt-file but also in shelve-dbase
<ALGNAME>_tistos.db.
If really large statistics is required there are some utilities to merge the
information from these <ALGNAME>_tistos.db together, e.g. on the output of Ganga jobs.
{% enddiscussion %}
{% challenge "Challenge" %}
- Add
decisions-function for your previous Bender module with n-tuples.- (Do not forget to instrument the
initializemethod)
- (Do not forget to instrument the
- Run it and observe the output summary table
- Identify the relevant
L0-TOS,Hlt1-TOSandHlt2-TOSlines for your decay
Now, when we have the lists of the relevant lines, one wants to add infrommation about the decisions of these lines into n-tuple/tree.
The method tistos is out friend here. To use this method one needs to instrument initialize-method:
class TisTosTuple(Algo):
"""Enhanced functionality for n-tuples
"""
def initialize ( self ) :
sc = Algo.initialize ( self ) ## initialize the base class
if sc.isFailure() : return sc
#
## container to collect trigger information, e.g. list of fired lines
#
triggers = {}
triggers ['psi'] = {} ## slot to keep information for J/psi
#
## the lines to be investigated in details
#
lines = {}
lines [ "psi" ] = {} ## trigger lines for J/psi
## six mandatory keys:
lines [ "psi" ][ 'L0TOS' ] = 'L0(DiMuon|Muon)Decision'
lines [ "psi" ][ 'L0TIS' ] = 'L0(Hadron|DiMuon|Muon|Electron|Photon)Decision'
lines [ "psi" ][ 'Hlt1TOS' ] = 'Hlt1(DiMuon|TrackMuon).*Decision'
lines [ "psi" ][ 'Hlt1TIS' ] = 'Hlt1(DiMuon|SingleMuon|Track).*Decision'
lines [ "psi" ][ 'Hlt2TOS' ] = 'Hlt2DiMuon.*Decision'
lines [ "psi" ][ 'Hlt2TIS' ] = 'Hlt2(Charm|Topo|DiMuon|Single).*Decision'
sc = self.tisTos_initialize ( triggers , lines )
if sc.isFailure() : return sc
return SUCCESSHere one defines six regex-patterns that describe the six sets of triggers lines:
L0, Hlt1, Hlt2 vs TIS, TOS. These expressions are coded according to the infomration,
obtainer earlier. The next step is rather trivial: in analyse method one need to invoke
the method tistos for the given particle, J/psi in our case:
def analyse( self ) :
"""The main 'analysis' method"""
...
tup = self.nTuple('MyTuple')
for b in particles :
psi =b(1) ## the first daughter: J/psi
...
## fill n-tuple with TISTOS information for J/psi
self.tisTos (
psi , ## particle
tup , ## n-tuple
'psi_' , ## prefix for variable name in n-tuple
self.lines['psi'] , ## trigger lines to be inspected
verbose = True ## good ooption for those who hates bit-wise operations
)
tup.write()
...
return SUCCESSThis code adds several variables into n-tuple/tree, see log-file or use help(Algo.tistos).
Also TisTos-information for global and physics triggers is added.
{% discussion "In details," %}
The fragment from log-file:
# BenderTools.TisTos INFO tisTos: Fill TisTos information into n-tuple
#
# # for d in particles :
# # self.tisTos ( d ,
# # tup ,
# # 'd0_' ,
# # self.lines ['D0'] ,
# # self.l0tistos ,
# # self.l1tistos ,
# # self.l2tistos )
#
# ``lines'' here is a dictionary of lines (or regex-patterns) with
# following keys:
# - L0TOS
# - L0TIS
# - Hlt1TOS
# - Hlt1TIS
# - Hlt2TOS
# - Hlt2TIS
#
# e.g.
#
# # lines = {}
# # lines ['L0TOS' ] = 'L0HadronDecision'
# # lines ['L0TIS' ] = 'L0(Hadron|Muon|DiMuon)Decision'
# # lines ['Hlt1TOS'] = ...
# # lines ['Hlt1TIS'] = 'Hlt1Topo.*Decision'
# # lines ['Hlt2TOS'] = ...
# # lines ['Hlt2TIS'] = ...
#
# Technically it is useful to keep it as ``per-particle-type'' dictionary
#
# # def initialize ( self ) :
# # ...
# # self.lines = {}
# # self.lines ['B' ] = {}
# # self.lines ['B' ]['L0TOS'] = ...
# # self.lines ['B' ]['L0TOS'] = ...
# # ...
# # self.lines ['psi'] = {}
# # self.lines ['psi']['L0TOS'] = ...
# # ...
# # return SUCCESS
#
# # def analyse ( self ) :
#
# # particles = ...
#
# # for B in particles :
# # self.tisTos ( B ,
# # tup ,
# # 'B0_' ,
# # self.lines['B'] ,
# # self.l0tistos ,
# # self.l1tistos ,
# # self.l2tistos )
#
# # ...
# # return SUCCESS
#
# The function adds few columns to n-tuple, the most important are
# - ``<label>l0tos'' that corresponds to 'L0-TOS'
# - ``<label>l0tis'' that corresponds to 'L0-TIS'
# - ``<label>l1tos'' that corresponds to 'Hlt1-TOS'
# - ``<label>l1tis'' that corresponds to 'Hlt1-TIS'
# - ``<label>l2tos'' that corresponds to 'Hlt2-TOS'
# - ``<label>l2tis'' that corresponds to 'Hlt2-TIS'
# Additionally information for five predefined lists is stored:
# - ``<label>l0phys'' : L0-physics lines,
# - ``<label>l1phys'' : Hlt1-physics lines (routing bit #46)
# - ``<label>l2phys'' : Hlt2-physics lines (routing bit #77)
# - ``<label>l1glob'' : Hlt1Global decision
# - ``<label>l2glob'' : Hlt2Global decision
#
# The stored value is unsigned short, a bit-representaion of ITisTos::TisTosTob object
# - see ITisTos
# - see ITisTos::TisTosTob
#
# Later, in processing of TTree one can use these flags as :
#
# >>> tree = ...
# >>> tree.Draw('M' ) ## no trigger requirements
#
# >>> tree.Draw('M', '(l0tos&2)==2') ## require L0-tos with respect to the list of 'L0TOS'-lines
# >>> tree.Draw('M', '(l1tos&2)==2') ## require Hlt1-tos with respect to the list of 'Hlt1TOS'-lines
# >>> tree.Draw('M', '(l2tos&2)==2') ## require Hlt2-tos with respect to the list of 'Hlt2TOS'-lines
#
# >>> tree.Draw('M', '(l0tis&4)==4') ## require L0-tis with respect to the list of 'L0TIS'-lines
# >>> tree.Draw('M', '(l1tis&4)==4') ## require Hlt1-tis with respect to the list of 'Hlt1TIS'-lines
# >>> tree.Draw('M', '(l2tis&4)==4') ## require Hlt2-tis with respect to the list of 'Hlt2TIS'-lines
#
# >>> tree.Draw('M', '(l0tos&3)==3') ## require L0-tus with respect to the list of 'L0TOS'-lines
# >>> tree.Draw('M', '(l1tos&3)==3') ## require Hlt1-tus with respect to the list of 'Hlt1TOS'-lines
# >>> tree.Draw('M', '(l2tos&3)==3') ## require Hlt2-tus with respect to the list of 'Hlt2TOS'-lines
#
# >>> tree.Draw('M', '(l0tos&1)==1') ## require L0-tps with respect to the list of 'L0TOS'-lines
# >>> tree.Draw('M', '(l1tos&1)==1') ## require Hlt1-tps with respect to the list of 'Hlt1TOS'-lines
# >>> tree.Draw('M', '(l2tos&1)==1') ## require Hlt2-tps with respect to the list of 'Hlt2TOS'-lines
#
# >>> tree.Draw('M', '(l0phys&2)==2') ## require L0-tos with respect to L0-physics lines
# >>> tree.Draw('M', '(l0phys&4)==4') ## require L0-tis with respect to L0-physics lines
#
# One can avoid bit-wise operations using ``verbose=True'' flag.
# In this case one gets following boolean variables in n-tuple:
# - ``<labeltag>_tos'' Trigger On Signal
# - ``<labeltag>_tis'' Trigger Independently on Signal
# - ``<labeltag>_tps'' Trigger Partially on Signal
# - ``<labeltag>_tus'' Trigger Used Signal (== TOS || TPS )
# - ``<labeltag>_dec'' Trigger decision
#
# where ``<labeltag>'' is
# - ``<label>l0tos''
# - ``<label>l0tis''
# - ``<label>l0phys'
# - ``<label>l1tos''
# - ``<label>l1tis''
# - ``<label>l1phys'
# - ``<label>l1glob'
# - ``<label>l2tos''
# - ``<label>l2tis''
# - ``<label>l2phys'
# - ``<label>l2glob'
{% enddiscussion %}
{% challenge "Challenge" %}
- Add
tistos-function for your previous Bender module with n-tuples.- (Do not forget to instrument the
initializemethod)
- (Do not forget to instrument the
- Run it and observe new variables in n-tuple/tree
- Make a plot of B-mass for all candidates, and for candiated that are
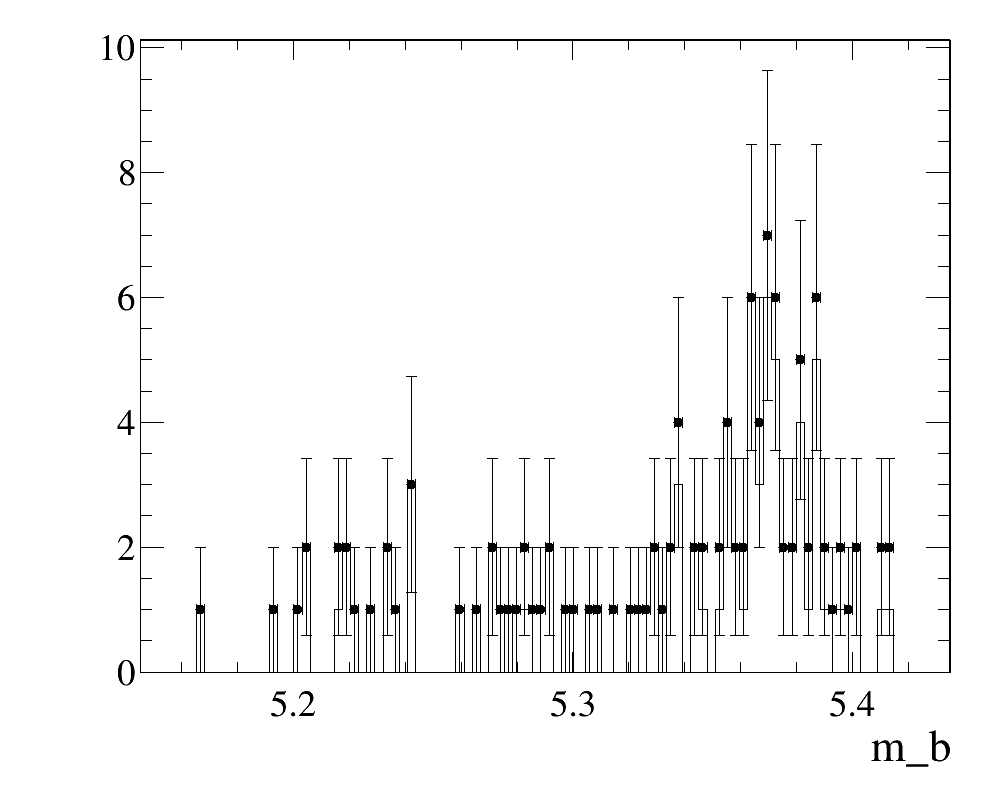
L0-TOS,Hlt-TOSandHlt2-TOSwith respect to the set of selected trigger lines. {% solution "Solution" %} The complete module is accessible here and the corresponsing log-file is here To make the corresponsing plot, e.g. start(i)python:
import ROOT
f = ROOT.TFile('TisTosTuples.root','READ')
t = f.Get('TisTos/MyTuple')
t.Draw ( 'm_b' ,'' , 'e1' )
t.Draw ( 'm_b' , ' psi_l0tos_tos && psi_l1tos_tos && psi_l2tos_tos' , 'same hist' )