Exploring ways to create a Python module that can let users use the XSPEC model library with minimal effort. The idea would then be that packages like Sherpa and 3ML could build on this package.
Well, that's the plan. I need people to actually try it out to see if it is useful and worth moving forward.
The home page for this module is xspec-models-cxc.
This is released under the GNU GPL version 3 as this is built on code developed for Sherpa and the CIAO contrib packages.
You need to have the XSPEC model library installed. The easiest way to
do this is to actually build and install
XSPEC directly, but it
should also work if you build just the XSPEC models library with the
--enable-xs-models-only flag. An alternative is to use the
CXC-provided xspec-modelsonly conda package that comes as part of
the CXC CIAO distribution.
Supported versions of XSPEC: 12.12.1 to 12.14.1.
Newer versions may work, but there's no guarantee since HEASOFT does change the build from time to time. Support for the older versions is not guaranteed either!
You need to have the HEADAS environment variable set up and probably
have also sourced the $HEADAS/headas-init.sh or
$HEADAS/headas-init.csh script. I also strongly suggest using
a new venv or conda environment!
The code will guess whether to use g++ or clang++. This choice will be over-ridden by setting the CXX environment variable (useful for cases where XSPEC was built with clang but you also have gcc installed, as the install defaults to g++ in this case).
Then you can either
% pip install xspec_models_cxc[test] --verbose
(the --verbose is in case there's an error, as the default information
you get from pip on a failue is generally less-than useful), or you
can try
% git clone https://github.com/cxcsds/xspec-models-cxc
% cd xspec-models-cxc
% pip install .[test] --verbose
The build requires both pybind11 and parse-xspec but they will be installed automatically if needed. Neither is required to use the compiled module.
Testing is done with (the actual output depends on the version of XSPEC installed and the version of this module):
% pytest
============================= test session starts ==============================
platform linux -- Python 3.12.6, pytest-8.3.3, pluggy-1.5.0
rootdir: /home/dburke/sherpa/xspec-models-cxc
configfile: pyproject.toml
collected 788 items
src/xspec_models_cxc/tests/test_basic.py ............................... [ 3%]
........................................................................ [ 13%]
..................................s..................................... [ 22%]
......s................................................................. [ 31%]
........................................................................ [ 40%]
................s...........................................s........... [ 49%]
........................................................................ [ 58%]
........................................................................ [ 67%]
.....................................ss....s.. [ 73%]
src/xspec_models_cxc/tests/test_realarray.py ........................... [ 77%]
........................................................................ [ 86%]
........................................................................ [ 95%]
.................................... [100%]
================== 781 passed, 7 skipped in 144.97s (0:02:24) ==================
delta 17.767712895533830
Ldisc 1.3800000596009081E+046 erg s^-1
RBLR 2506.2608817039027 Rg 3.7148351461236269E+017 cm
RIR 62656.522903742269 Rg 9.2870879929498337E+018 cm
mdot>=0.01, so jet = SSC + EC
gcool 7.2043275554242925
log Pr 45.3022881 log Pb 45.1834221
log Pe 44.5943985 log Pp 46.9754486
log Pj 46.9931221
idre: initializing data tables, please wait...
Ref.: Dovciak M., Karas V. & Yaqoob T.
ApJS July 2004, Volume 153, Issue 1, pp. 205-221
------------------------------------------------------
...initializing finished
-------setup for qsosed------
Gamma_warm= 2.5000000000000000 kTe_warm= 0.20000000000000001
Gamma_hard=internal calculation kTe_hot= 100.00000000000000
albedo= 0.29999999999999999
Rwarm/Rhot= 2.0000000000000000
Htmax= 100.00000000000000
------hot compton-----
Gamma_hot= 1.9943788473581052 Rhot= 14.262535224556565
T(Rhot)nt= 115584.83219658821 Tseed= 216069.57323613123
Ldis,hot/Ledd= 2.0029989017115401E-002 Lhot/Ledd= 2.4198583118457476E-002
------warm compton-----
T(Rw)= 80093.592123168855 Rwarm= 28.525070449113130
------Rout-----
rout(rsg)= 1288.8911460759962
tout= 5592.6110556431468
-----------
delta 17.767712895533830
Ldisc 1.3800000596009081E+046 erg s^-1
RBLR 2506.2608817039027 Rg 3.7148351461236269E+017 cm
RIR 62656.522903742269 Rg 9.2870879929498337E+018 cm
mdot>=0.01, so jet = SSC + EC
gcool 7.2043275554242925
log Pr 45.3022881 log Pb 45.1834221
log Pe 44.5943985 log Pp 46.9754486
log Pj 46.9931221
-------setup for qsosed------
Gamma_warm= 2.5000000000000000 kTe_warm= 0.20000000000000001
Gamma_hard=internal calculation kTe_hot= 100.00000000000000
albedo= 0.29999999999999999
Rwarm/Rhot= 2.0000000000000000
Htmax= 100.00000000000000
------hot compton-----
Gamma_hot= 1.9943788473581052 Rhot= 14.262535224556565
T(Rhot)nt= 115584.83219658821 Tseed= 216069.57323613123
Ldis,hot/Ledd= 2.0029989017115401E-002 Lhot/Ledd= 2.4198583118457476E-002
------warm compton-----
T(Rw)= 80093.592123168855 Rwarm= 28.525070449113130
------Rout-----
rout(rsg)= 1288.8911460759962
tout= 5592.6110556431468
-----------
ISMabs: ISM absorption model Version1.2
Gatuzz, Garcia, Kallman, Mendoza, & Gorczyca (2014)
Note: Default column densities are given
according to Grevesse, N. & Sauval (1998)
assuming N_H = 1.E21 cm^-2
idre: initializing data tables, please wait...
Ref.: Dovciak M., Karas V. & Yaqoob T.
ApJS July 2004, Volume 153, Issue 1, pp. 205-221
------------------------------------------------------
...initializing finished
The $HEADAS/../spectral/manager/model.dat file is used to determine
what models are available, and their parameters.
Number of models: 293
| Type | Total | Supported |
|---|---|---|
| additive | 203 | 203 |
| multiplicative | 67 | 67 |
| convolution | 23 | 23 |
| acn | 1 | 0 |
| Type | Total | Supported |
|---|---|---|
| C++ | 197 | 196 |
| C | 8 | 8 |
| FORTRAN sp | 86 | 86 |
| FORTRAN dp | 3 | 3 |
Number skipped: 1
The pileup model is unsupported (as it uses the "acn" model type, as are
any of the mixing models, which we do not even try to support).
The interface mirrors that used by the XSPEC local model interface, but tweaked to support Python and NumPy.
As shown below, each model is represented by a function in the
xspec_models_cxc module which takes an energies and pars
argument, and returns a NumPy array (there are some tweaks, such as
the optional spectrum argument for those models using XFLT data,
convolution models needing more arguments, and different ways to
evaluate the model). More information is provided in the Evaluating
Models
section below.
Here's a quick run through, which is available as scripts/example.py. The Examples section below has more details.
import numpy as np
from matplotlib import pyplot as plt
import xspec_models_cxc as x
x.chatter(0) # Hide the screen messages
vxspec = x.get_version()
print(f"XSPEC version: {vxspec}")
print(f"Module version: {x.__version__}")
def add_version():
plt.text(0.98, 0.98, f"XSPEC {vxspec}",
transform=plt.gcf().transFigure,
verticalalignment="top",
horizontalalignment="right")
plt.text(0.02, 0.98, f"Module {x.__version__}",
transform=plt.gcf().transFigure,
verticalalignment="top",
horizontalalignment="left")
egrid = np.arange(0.1, 20, 0.01)
emid = (egrid[:-1] + egrid[1:]) / 2
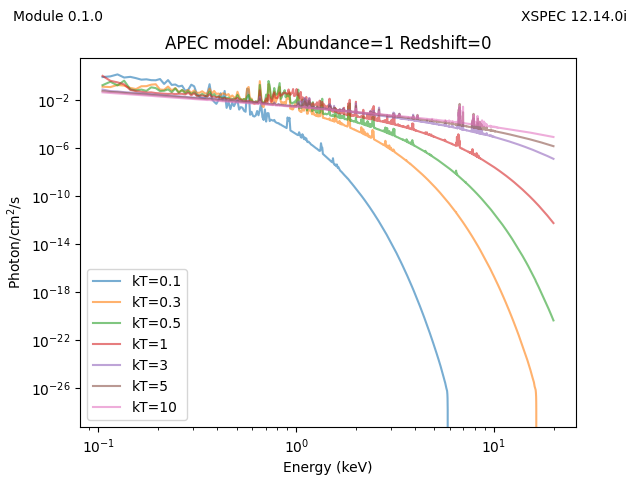
for kT in [0.1, 0.3, 0.5, 1, 3, 5, 10]:
y = x.apec(energies=egrid, pars=[kT, 1, 0])
plt.plot(emid, y, label=f'kT={kT}', alpha=0.6)
plt.xscale('log')
plt.yscale('log')
plt.legend()
plt.xlabel('Energy (keV)')
plt.ylabel('Photon/cm$^2$/s')
plt.title('APEC model: Abundance=1 Redshift=0')
add_version()
plt.savefig('example-additive.png')
plt.clf()
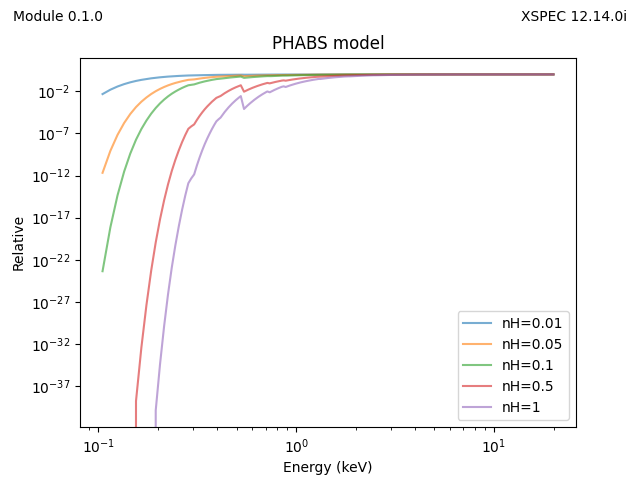
for nH in [0.01, 0.05, 0.1, 0.5, 1]:
y = x.phabs(energies=egrid, pars=[nH])
plt.plot(emid, y, label=f'nH={nH}', alpha=0.6)
plt.xscale('log')
plt.yscale('log')
plt.legend()
plt.xlabel('Energy (keV)')
plt.ylabel('Relative')
plt.title('PHABS model')
add_version()
plt.savefig('example-multiplicative.png')
plt.clf()
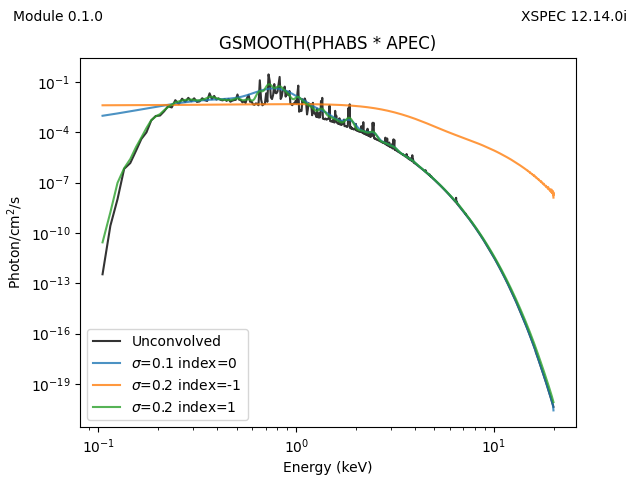
model = x.phabs(energies=egrid, pars=[0.05]) * x.apec(energies=egrid, pars=[0.5, 1, 0])
plt.plot(emid, model, label='Unconvolved', c='k', alpha=0.8)
for pars in [[0.1, 0], [0.2, -1], [0.2, 1]]:
# the model argument gets over-written by gsmooth, hence the copy
y = x.gsmooth(energies=egrid, pars=pars, model=model.copy())
plt.plot(emid, y, label=rf'$\sigma$={pars[0]} index={pars[1]}', alpha=0.8)
plt.xscale('log')
plt.yscale('log')
plt.legend()
plt.xlabel('Energy (keV)')
plt.ylabel('Photon/cm$^2$/s')
plt.title('GSMOOTH(PHABS * APEC)')
add_version()
plt.savefig('example-convolution.png')The screen output is just
XSPEC version: 12.14.1d
Module version: 0.1.0
and the plots are
The info() and list_models() routines give information on the
supported models.
>>> import xspec_models_cxc as x
>>> x.list_models()
['SSS_ice', 'TBabs', 'TBfeo', 'TBgas', 'TBgrain', 'TBpcf', ...
... 'zvphabs', 'zwabs', 'zwndabs', 'zxipab', 'zxipcf']
>>> x.list_models(modeltype=x.ModelType.Con)
['cflux', 'clumin', 'cpflux', 'gsmooth', 'ireflect', 'kdblur', 'kdblur2', 'kerrconv', 'kyconv', 'lsmooth', 'partcov', 'rdblur', 'reflect', 'rfxconv', 'rgsxsrc', 'simpl', 'thcomp', 'vashift', 'vmshift', 'xilconv', 'zashift', 'zmshift']
>>> x.list_models(modeltype=x.ModelType.Con, language=x.LanguageStyle.F77Style4)
['kyconv', 'rgsxsrc', 'thcomp']
>>> x.list_models(language=x.LanguageStyle.F77Style8)
['ismabs', 'ismdust', 'olivineabs']
>>> x.info('apec')
XSPECModel(modeltype=<ModelType.Add: 1>, name='apec', funcname='apec', language=<LanguageStyle.CppStyle8: 1>, elo=0.0, ehi=1e+20, parameters=[XSPECParameter(paramtype=<ParamType.Default: 1>, name='kT', default=1.0, units='keV', frozen=False, softmin=0.008, softmax=64.0, hardmin=0.008, hardmax=64.0, delta=0.01), XSPECParameter(paramtype=<ParamType.Default: 1>, name='Abundanc', default=1.0, units=None, frozen=True, softmin=0.0, softmax=5.0, hardmin=0.0, hardmax=5.0, delta=0.001), XSPECParameter(paramtype=<ParamType.Default: 1>, name='Redshift', default=0.0, units=None, frozen=True, softmin=-0.999, softmax=10.0, hardmin=-0.999, hardmax=10.0, delta=0.01)], use_errors=False, can_cache=True)
>>> x.info('TBabs')
XSPECModel(modeltype=<ModelType.Mul: 2>, name='TBabs', funcname='tbabs', language=<LanguageStyle.CppStyle8: 1>, elo=0.03, ehi=1e+20, parameters=[XSPECParameter(paramtype=<ParamType.Default: 1>, name='nH', default=1.0, units='10^22', frozen=False, softmin=0.0, softmax=100000.0, hardmin=0.0, hardmax=1000000.0, delta=0.001)], use_errors=False, can_cache=True)
>>> x.info('zxipab')
XSPECModel(modeltype=<ModelType.Mul: 2>, name='zxipab', funcname='zxipab', language=<LanguageStyle.F77Style4: 3>, elo=0.01, ehi=1e+20, parameters=[XSPECParameter(paramtype=<ParamType.Default: 1>, name='nHmin', default=0.01, units='10^22', frozen=False, softmin=1e-07, softmax=1000.0, hardmin=1e-07, hardmax=1000000.0, delta=0.01), XSPECParameter(paramtype=<ParamType.Default: 1>, name='nHmax', default=10.0, units='10^22', frozen=False, softmin=1e-07, softmax=1000.0, hardmin=1e-07, hardmax=1000000.0, delta=0.01), XSPECParameter(paramtype=<ParamType.Default: 1>, name='beta', default=0.0, units=None, frozen=False, softmin=-10.0, softmax=10.0, hardmin=-10.0, hardmax=10.0, delta=0.01), XSPECParameter(paramtype=<ParamType.Default: 1>, name='log_xi', default=3.0, units=None, frozen=False, softmin=-3.0, softmax=6.0, hardmin=-3.0, hardmax=6.0, delta=0.01), XSPECParameter(paramtype=<ParamType.Default: 1>, name='redshift', default=0.0, units=None, frozen=True, softmin=0.0, softmax=10.0, hardmin=0.0, hardmax=10.0, delta=0.01)], use_errors=False, can_cache=True)
>>> x.info('smaug')
XSPECModel(modeltype=<ModelType.Add: 1>, name='smaug', funcname='xsmaug', language=<LanguageStyle.CStyle8: 2>, elo=0.0, ehi=1e+20, parameters=[XSPECParameter(paramtype=<ParamType.Default: 1>, name='kT_cc', default=1.0, units='keV', frozen=False, softmin=0.1, softmax=10.0, hardmin=0.08, hardmax=100.0, delta=0.01), XSPECParameter(paramtype=<ParamType.Default: 1>, name='kT_dt',
default=1.0, units='keV', frozen=False, softmin=0.0, softmax=10.0, hardmin=0.0, hardmax=100.0, delta=0.01), XSPECParameter(paramtype=<ParamType.Default: 1>, name='kT_ix', default=0.0, units=None, frozen=True, softmin=0.0, softmax=10.0, hardmin=0.0, hardmax=10.0, delta=0.001), XSPECParameter(paramtype=<ParamType.Default: 1>, name='kT_ir', default=0.1, units='Mpc', frozen=True, softmin=0.0001, softmax=1.0, hardmin=0.0001, hardmax=1.0, delta=0.001), XSPECParameter(paramtype=<ParamType.Default: 1>, name='kT_cx', default=0.5, units=None, frozen=False, softmin=0.0, softmax=10.0, hardmin=0.0, hardmax=10.0, delta=0.001), XSPECParameter(paramtype=<ParamType.Default: 1>, name='kT_cr', default=0.1, units='Mpc', frozen=False, softmin=0.0001, softmax=10.0, hardmin=0.0001, hardmax=20.0, delta=0.01), XSPECParameter(paramtype=<ParamType.Default: 1>, name='kT_tx', default=0.0, units=None, frozen=True, softmin=0.0, softmax=10.0, hardmin=0.0, hardmax=10.0, delta=0.001), XSPECParameter(paramtype=<ParamType.Default: 1>, name='kT_tr', default=0.5, units='Mpc', frozen=True, softmin=0.0001, softmax=1.0, hardmin=0.0001, hardmax=3.0, delta=0.01), XSPECParameter(paramtype=<ParamType.Default: 1>, name='nH_cc', default=1.0, units='cm**-3', frozen=True, softmin=1e-06, softmax=3.0, hardmin=1e-06, hardmax=3.0, delta=0.01), XSPECParameter(paramtype=<ParamType.Default: 1>, name='nH_ff', default=1.0, units=None, frozen=True, softmin=0.0, softmax=1.0, hardmin=0.0, hardmax=1.0, delta=0.01), XSPECParameter(paramtype=<ParamType.Default: 1>, name='nH_cx', default=0.5, units=None, frozen=False, softmin=0.0, softmax=10.0, hardmin=0.0, hardmax=10.0, delta=0.001), XSPECParameter(paramtype=<ParamType.Default: 1>, name='nH_cr', default=0.1, units='Mpc', frozen=False, softmin=0.0001, softmax=1.0, hardmin=0.0001, hardmax=2.0, delta=0.01), XSPECParameter(paramtype=<ParamType.Default: 1>, name='nH_gx', default=0.0, units=None, frozen=True, softmin=0.0, softmax=10.0, hardmin=0.0, hardmax=10.0, delta=0.001), XSPECParameter(paramtype=<ParamType.Default: 1>, name='nH_gr', default=0.002, units='Mpc', frozen=True, softmin=0.0001, softmax=10.0, hardmin=0.0001,
hardmax=20.0, delta=0.001), XSPECParameter(paramtype=<ParamType.Default: 1>, name='Ab_cc', default=1.0, units='solar', frozen=True, softmin=0.0, softmax=3.0, hardmin=0.0, hardmax=5.0, delta=0.01), XSPECParameter(paramtype=<ParamType.Default: 1>, name='Ab_xx', default=0.0, units=None, frozen=True, softmin=0.0, softmax=10.0, hardmin=0.0, hardmax=10.0, delta=0.001), XSPECParameter(paramtype=<ParamType.Default: 1>, name='Ab_rr', default=0.1, units='Mpc', frozen=True, softmin=0.0001, softmax=1.0, hardmin=0.0001, hardmax=1.0, delta=0.01), XSPECParameter(paramtype=<ParamType.Default: 1>, name='redshift', default=0.01, units=None, frozen=True, softmin=0.0001, softmax=10.0, hardmin=0.0001, hardmax=10.0, delta=1.0), XSPECParameter(paramtype=<ParamType.Default: 1>, name='meshpts', default=10.0, units=None, frozen=True, softmin=1.0, softmax=10000.0, hardmin=1.0, hardmax=10000.0, delta=1.0), XSPECParameter(paramtype=<ParamType.Default: 1>, name='rcutoff', default=2.0, units='Mpc', frozen=True, softmin=1.0, softmax=3.0, hardmin=1.0, hardmax=3.0, delta=0.01), XSPECParameter(paramtype=<ParamType.Default: 1>, name='mode', default=1.0, units=None, frozen=True, softmin=0.0, softmax=2.0, hardmin=0.0, hardmax=2.0, delta=1.0), XSPECParameter(paramtype=<ParamType.Default: 1>, name='itype', default=2.0, units=None, frozen=True, softmin=1.0, softmax=4.0, hardmin=1.0, hardmax=4.0, delta=1.0)], use_errors=False, can_cache=False)
>>> m = x.info('smaug')
>>> for p in m.parameters:
... print(f' {p.name:10s} = {p.default:5g} units={p.units} frozen={p.frozen} range: {p.hardmin}-{p.hardmax}')
...
kT_cc = 1 units=keV frozen=False range: 0.08-100.0
kT_dt = 1 units=keV frozen=False range: 0.0-100.0
kT_ix = 0 units=None frozen=True range: 0.0-10.0
kT_ir = 0.1 units=Mpc frozen=True range: 0.0001-1.0
kT_cx = 0.5 units=None frozen=False range: 0.0-10.0
kT_cr = 0.1 units=Mpc frozen=False range: 0.0001-20.0
kT_tx = 0 units=None frozen=True range: 0.0-10.0
kT_tr = 0.5 units=Mpc frozen=True range: 0.0001-3.0
nH_cc = 1 units=cm**-3 frozen=True range: 1e-06-3.0
nH_ff = 1 units=None frozen=True range: 0.0-1.0
nH_cx = 0.5 units=None frozen=False range: 0.0-10.0
nH_cr = 0.1 units=Mpc frozen=False range: 0.0001-2.0
nH_gx = 0 units=None frozen=True range: 0.0-10.0
nH_gr = 0.002 units=Mpc frozen=True range: 0.0001-20.0
Ab_cc = 1 units=solar frozen=True range: 0.0-5.0
Ab_xx = 0 units=None frozen=True range: 0.0-10.0
Ab_rr = 0.1 units=Mpc frozen=True range: 0.0001-1.0
redshift = 0.01 units=None frozen=True range: 0.0001-10.0
meshpts = 10 units=None frozen=True range: 1.0-10000.0
rcutoff = 2 units=Mpc frozen=True range: 1.0-3.0
mode = 1 units=None frozen=True range: 0.0-2.0
itype = 2 units=None frozen=True range: 1.0-4.0
New in version 0.0.20 is support for XSPEC table models. This support is rather limited as it only allows you toe valuate the table model and does not provide any access to the metadata in the model (which determines whether it is an atable or mtable model [unfortunately there is no flag to say it's an etable model], whether to add a redshift parameter, what the parameters are, what the default and parameter ranges are, ...).
Models are evaluated with the tableModel function.
Unfortunately, because of the way that XSPEC provides access to the table models, it will not work with XSPEC 12.12.0 (or, rather, it is not clear how best to support it in this version whilst also supporting XSPEC 12.12.1, so I've dropped the 12.12.0 support for now).
The XSPEC model library is automatically initalized when the first call
is made, not when the module is loaded. The init function provided
in version 0.0.5 and earlier is no-longer provided.
>>> import xspec_models_cxc as x
>>> x.__version__
'0.0.29'
>>> help(x)
Help on package xspec_models_cxc:
NAME
xspec_models_cxc - Experiment with XSPEC models.
DESCRIPTION
The XSPEC model library is automatically initialized when the first
call is made, and not when the module is loaded.
There are three types of symbols in this package:
1. model functions, such as `apec` and `TBabs`, and `tableModel` for
table models.
2. routines that set or get values such as the abundance
table (`abundance`), cross-section table (`cross_sections`),
cosmology (`cosmology`), and the chatter level (`chatter`).
3. routines about the models: `info` and `list_models`.
Examples
--------
What version of XSPEC is being used?
>>> import xspec_models_cxc as x
>>> x.get_version()
'12.14.1'
What models are supported (the actual list depends on the
version of XSPEC the code was compiled against)?
>>> import xspec_models_cxc as x
>>> x.list_models()
['SSS_ice', 'TBabs', 'TBfeo', ..., 'zwndabs', 'zxipab', 'zxipcf']
Evaluate the multiplication of phabs and apec models, with their
default parameter ranges, on the grid from 0.1 to 10 keV with a bin
size of 1 eV. Note that the return value has one less element than the
grid, because it is evaluated over the range egrid[0]-egrid[1],
egrid[1]-egrid[2], ..., egrid[-2]-egrid[-1] - which for this case is
0.100-0.101, 0.101-0.102, ..., 9.998-9.999 keV.
>>> import numpy as np
>>> from matplotlib import pyplot as plt
>>> import xspec_models_cxc as x
>>> egrid = np.arange(0.1, 10, 0.001)
>>> apec = x.info('apec')
>>> phabs = x.info('phabs')
>>> pars_apec = [p.default for p in apec.parameters]
>>> pars_phabs = [p.default for p in phabs.parameters]
>>> yapec = x.apec(energies=egrid, pars=pars_apec)
>>> yphabs = x.phabs(energies=egrid, pars=pars_phabs)
>>> ymodel = yphabs * yapec
>>> emid = (egrid[:-1] + egrid[1:]) / 2
>>> plt.plot(emid, ymodel, label='phabs * apec')
>>> plt.yscale('log')
>>> plt.ylim(1e-9, 0.01)
>>> plt.legend()
>>> plt.xlabel('Energy (keV)')
>>> plt.ylabel('Photon/cm$^2$/s')
We can include a convolution component - in this case the kdblur model
- even if it is physically unrealistic. The two differences here is
that the model requires the data to be convolved to be sent in as the
`model` argument, and that this array is changed by the routine (in
the same way that the out parameter works for NumPy ufunc
routines). The convolution models also return the value so we could
have said
out = x.kdblur(ebergies=.., pars=.., model=ymodel.copy())
which would keep the original model values (there is actualy a subtly
in that the `model` argument must be sent the correct datatype for the
convolution model - so either `np.float64` or `np.float32` - otherwise
it will not be changed).
>>> kdblur = x.info('kdblur')
>>> pars_kdblur = [p.default for p in kdblur.parameters]
>>> x.kdblur(energies=egrid, pars=pars_kdblur, model=ymodel)
>>> plt.plot(emid, ymodel, alpha=0.8, label='Convolved')
>>> plt.legend()
XSPEC table models [TableModel]_ are fun to work with, as you
1. need to read in the file to find out information on the model -
such as whether it's atable or mtable (but unfortunately there is
no header keyword to determine if it is an etable) - and the
parameter names, values, and ranges.
2. use the file name when evaluating the model along with some of
this metadata.
At the moment this module only supports the second part - calling the
models - and it is left to the user to find the other information out.
In this example the ``RCS.mod`` table model, which has three
parameters, does not add a redshift parameter, and is an "atable"
model (i.e. additive):
% dmlist "RCS.mod[cols name, initial]" data,clean
# NAME INITIAL
tau 1.0
beta 0.10000000149012
T 0.10000000149012
% dmkeypar xspec-tablemodel-RCS.mod"[primary]" redshift echo+
0
% dmkeypar "xspec-tablemodel-RCS.mod[primary]" addmodel echo+
1
This can then be used with `tableModel` in a similar manner to the
other models, apart from requiring `table` and `table_type` arguments:
>>> infile = 'RCS.mod'
>>> pars = [1, 0.1, 0.1]
>>> egrid = np.arange(0.1, 10, 0.01)
>>> y = x.tableModel(table=infile, table_type="add", energies=egrid, pars=pars)
Note that it is very easy to make the table model code crash the
system, such as by sending in not enough parameters or setting a
parameter outside its hard limits:
>>> x.tableModel(infile, "add", pars=[1, 2], energies=egrid)
Segmentation fault (core dumped)
References
----------
.. [TableModel] https://heasarc.gsfc.nasa.gov/docs/heasarc/ofwg/docs/general/ogip_92_009/ogip_92_009.html
PACKAGE CONTENTS
_compiled
CLASSES
builtins.object
XSPECModel
XSPECParameter
enum.Enum(builtins.object)
LanguageStyle
ModelType
ParamType
...
FUNCTIONS
SSS_ice(...) method of builtins.PyCapsule instance
SSS_ice(*args, **kwargs)
Overloaded function.
1. SSS_ice(pars: numpy.ndarray[numpy.float32], energies: numpy.ndarray[numpy.float32], spectrum: int = 1) -> numpy.ndarray[numpy.float32]
The XSPEC multiplicative SSS_ice model (1 parameter).
2. SSS_ice(pars: numpy.ndarray[numpy.float32], energies: numpy.ndarray[numpy.float32], out: numpy.ndarray[numpy.float32], spectrum: int = 1) -> numpy.ndarray[
numpy.float32]
The XSPEC multiplicative SSS_ice model (1 parameter); inplace.
TBabs(...) method of builtins.PyCapsule instance
TBabs(*args, **kwargs)
Overloaded function.
1. TBabs(pars: numpy.ndarray[numpy.float64], energies: numpy.ndarray[numpy.float64], spectrum: int = 1, initStr: str = '') -> numpy.ndarray[numpy.float64]
The XSPEC multiplicative TBabs model (1 parameter).
2. TBabs(pars: numpy.ndarray[numpy.float64], energies: numpy.ndarray[numpy.float64], out: numpy.ndarray[numpy.float64], spectrum: int = 1, initStr: str = '') -> numpy.ndarray[numpy.float64]
The XSPEC multiplicative TBabs model (1 parameter); inplace.
TBabs_(...) method of builtins.PyCapsule instance
TBabs_(pars: xspec_models_cxc._compiled.RealArray, energies: xspec_models_cxc._compiled.RealArray, out: xspec_models_cxc._compiled.RealArray, spectrum: int = 1, initStr: str = '') -> xspec_models_cxc._compiled.RealArray
The XSPEC multiplicative TBabs model (1 parameter); RealArray, inplace.
TBfeo(...) method of builtins.PyCapsule instance
...
DATA
List = typing.List
A generic version of list.
Optional = typing.Optional
Optional[X] is equivalent to Union[X, None].
Sequence = typing.Sequence
A generic version of collections.abc.Sequence.
numberElements = 30
VERSION
0.0.29
FILE
/some/long/path/to/xspec-models-cxc/xspec_models_cxc.__init__.py
Note that you can see the difference between a FORTRAN model such as
SSS_ice, which deals with single-precision floats, and C/C++ models
such as TBabs, which deal with double-precision floats. See the
EVALUATING
MODELS
section below.
With this we can do a few things:
>>> help(x.get_version)
Help on built-in function get_version in module xspec_models_cxc:
get_version(...) method of builtins.PyCapsule instance
get_version() -> str
The version of the XSPEC model library
>>> x.get_version()
'12.14.1'
>>> help(x.chatter)
Help on built-in function chatter in module xspec_models_cxc:
chatter(...) method of builtins.PyCapsule instance
chatter(*args, **kwargs)
Overloaded function.
1. chatter() -> int
Get the XSPEC chatter level.
2. chatter(chatter: int) -> None
Set the XSPEC chatter level.
>>> x.chatter()
10
>>> x.chatter(0)
>>> x.chatter()
0
>>> x.chatter(10)
>>> help(x.abundance)
Help on built-in function abundance in module xspec_models_cxc:
abundance(...) method of builtins.PyCapsule instance
abundance(*args, **kwargs)
Overloaded function.
1. abundance() -> str
Get the abundance-table setting.
2. abundance(table: str) -> None
Set the abundance-table setting.
>>> x.abundance()
'angr'
>>> x.abundance('lpgp')
Solar Abundance Vector set to lpgp: Lodders K., Palme H., Gail H.P., Landolt-Börnstein, New Series, vol VI/4B, pp 560–630 (2009) (Photospheric)
>>> x.abundance()
'lpgp'
>>> x.abundance('angr')
Solar Abundance Vector set to angr: Anders E. & Grevesse N. Geochimica et Cosmochimica Acta 53, 197 (1989)
It isn't clever enough to notice if you give it an unsupported abundance name.
>>> help(x.elementName)
Help on built-in function elementName in module xspec_models_cxc:
elementName(...) method of builtins.PyCapsule instance
elementName(z: int) -> str
Return the name of an element given the atomic number.
>>> x.elementName(17)
'Cl'
>>> help(x.elementAbundance)
Help on built-in function elementAbundance in module xspec_models_cxc:
elementAbundance(...) method of builtins.PyCapsule instance
elementAbundance(*args, **kwargs)
Overloaded function.
1. elementAbundance(name: str) -> float
Return the abundance setting for an element given the name.
2. elementAbundance(z: int) -> float
Return the abundance setting for an element given the atomic number.
>>> x.elementAbundance('Cl')
3.160000119351025e-07
>>> x.elementAbundance(17)
3.160000119351025e-07
Note that there's limited checking:
>>> >>> x.elementAbundance('Po')
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
KeyError: 'Po'
>>> x.elementAbundance(256)
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
IndexError: 256
>>> x.elementAbundance(0)
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
IndexError: 0
>>> x.elementAbundance(-4)
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
TypeError: elementAbundance(): incompatible function arguments. The following argument types are supported:
1. (name: str) -> float
2. (z: int) -> float
Invoked with: -4
Additive and multipicative models can either create a new output array on each call - such as
>>> y = x.apec(pars=pars, energies=egrid)
or they can re-use an output array (in a similar manner to the out
argument of NumPy routines like
np.cumsum):
>>> y = np.zeros(egrid.size - 1)
>>> yout = x.apec(pars=pars, energies=egrid, out=y)
>>> yout is y
True
The model.dat record for this model is
apec 3 0. 1.e20 C_apec add 0
kT keV 1. 0.008 0.008 64.0 64.0 .01
Abundanc " " 1. 0. 0. 5. 5. -0.001
Redshift " " 0. -0.999 -0.999 10. 10. -0.01
So, if we want to use the default parameters - that is, kT=1, Abundance=1, Redshift=0 - for the energy grid 0.1-0.2, 0.2-0.3, 0.3-0.4, and 0.4-0.5 we can say:
>>> import xspec_models_cxc as x
>>> help(x.apec)
Help on built-in function apec in module xspec_models_cxc:
apec(...) method of builtins.PyCapsule instance
apec(*args, **kwargs)
Overloaded function.
1. apec(pars: numpy.ndarray[numpy.float64], energies: numpy.ndarray[numpy.float64], spectrum: int = 1, initStr: str = '')
-> numpy.ndarray[numpy.float64]
The XSPEC additive apec model (3 parameters).
2. apec(pars: numpy.ndarray[numpy.float64], energies: numpy.ndarray[numpy.float64], out: numpy.ndarray[numpy.float64], spectrum: int = 1, initStr: str = '') -> numpy.ndarray[numpy.float64]
The XSPEC additive apec model (3 parameters); inplace.
>>> pars = [1, 1, 0]
>>> egrid = [0.1, 0.2, 0.3, 0.4, 0.5]
>>> x.apec(pars, egrid)
Reading APEC data from 3.0.9
array([2.10839183, 0.31196176, 0.22008776, 0.12295151])
>>>
We can see what dufference dropping the abundance to 0 makes:
>>> x.apec([1, 0, 0], egrid)
array([0.47038697, 0.21376409, 0.1247977 , 0.08182932])
Note that the return values have units of photons/cm^2/s as this is an XSPEC additive model.
The agnslim additive model is a FORTRAN model in 12.12.0:
agnslim 14 0.03 1.e20 agnslim add 0
mass solar 1e7 1.0 1.0 1.e10 1.e10 -.1
dist Mpc 100 0.01 0.01 1.e9 1.e9 -.01
logmdot " " 1. -10. -10. 3 3 0.01
astar " " 0. 0. 0. 0.998 0.998 -1
cosi " " 0.5 0.05 0.05 1. 1. -1
kTe_hot keV(-pl) 100.0 10 10 300 300 -1
kTe_warm keV(-sc) 0.2 0.1 0.1 0.5 0.5 1e-2
Gamma_hot " " 2.4 1.3 1.3 3 3. 0.01
Gamma_warm "(-disk)" 3.0 2 2 5. 10. 0.01
R_hot "Rg " 10.0 2.0 2.0 500 500 0.01
R_warm "Rg" 20.0 2 2 500 500 0.1
logrout "(-selfg) " -1.0 -3.0 -3.0 7.0 7.0 -1e-2
rin "" -1 -1 -1 100. 100. -1
redshift " " 0.0 0. 0. 5 5 -1
>>> help(x.agnslim)
Help on built-in function agnslim in module xspec_models_cxc:
agnslim(...) method of builtins.PyCapsule instance
agnslim(*args, **kwargs)
Overloaded function.
1. agnslim(pars: numpy.ndarray[numpy.float32], energies: numpy.ndarray[numpy.float32], spectrum: int = 1) -> numpy.ndarray[numpy.float32]
The XSPEC additive agnslim model (14 parameters).
2. agnslim(pars: numpy.ndarray[numpy.float32], energies: numpy.ndarray[numpy.float32], out: numpy.ndarray[numpy.float32],
spectrum: int = 1) -> numpy.ndarray[numpy.float32]
The XSPEC additive agnslim model (14 parameters); inplace.
>>> pars = [1e7, 100, 1, 0, 0.5, 100, 0.2, 2.4, 3, 10, 20, -1, -1, 0]
>>> egrid = np.arange(0.1, 11, 0.01)
>>> y = x.agnslim(pars, egrid)
>>> y
array([5.6430912e-01, 4.2761257e-01, 3.3259588e-01, ..., 2.6246285e-06,
2.6130140e-06, 2.6132632e-06], dtype=float32)
This is a C-style additive model:
bwcycl 12 0. 1.e20 c_beckerwolff add 0
Radius km 10 5 5 20 20 -1
Mass Solar 1.4 1 1 3 3 -1
csi " " 1.5 0.01 0.01 20 20 0.01
delta " " 1.8 0.01 0.01 20 20 0.01
B 1e12G 4 0.01 0.01 100 100 0.01
Mdot 1e17g/s 1 1e-6 1e-6 1e6 1e6 0.01
Te keV 5 0.1 0.1 100 100 0.01
r0 m 44 10 10 1000 1000 0.01
D kpc 5 1 1 20 20 -1
BBnorm " " 0 0 0 100 100 -1
CYCnorm " " 1 -1 -1 100 100 -1
FFnorm " " 1 -1 -1 100 100 -1
>>> help(x.bwcycl)
Help on built-in function bwcycl in module xspec_models_cxc:
bwcycl(...) method of builtins.PyCapsule instance
bwcycl(*args, **kwargs)
Overloaded function.
1. bwcycl(pars: numpy.ndarray[numpy.float64], energies: numpy.ndarray[numpy.float64], spectrum: int = 1, initStr: str = '') -> numpy.ndarray[numpy.float64]
The XSPEC additive bwcycl model (12 parameters).
2. bwcycl(pars: numpy.ndarray[numpy.float64], energies: numpy.ndarray[numpy.float64], out: numpy.ndarray[numpy.float64], spectrum: int = 1, initStr: str = '') -> numpy.ndarray[numpy.float64]
The XSPEC additive bwcycl model (12 parameters); inplace.
>>> pars = [10, 1.3, 1.5, 1.8, 4, 1, 5, 44, 5, 0, 1, 1]
>>> x.bwcycl(pars, [0.5, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56])
array([0.00030135, 0.00030085, 0.00030123, 0.00030297, 0.00030657,
0.00031248])
The TBabs is a multiplicative model:
TBabs 1 0.03 1.e20 C_tbabs mul 0
nH 10^22 1. 0. 0. 1E5 1E6 1E-3
With the default setting we don't expect much flux to get through for the selected energy range (~0.1 - 0.5 keV), but this increases as nH decreases by a magnitude or two:
>>> import numpy
>>> egrid = np.arange(0.1, 0.5, 0.05)
>>> x.abundance('wilm')
Solar Abundance Vector set to wilm: Wilms, J., Allen, A. & McCray, R. ApJ 542 914 (2000) (abundances are set to zero for those elements not included in the paper).
>>> help(x.TBabs)
Help on built-in function TBabs in module xspec_models_cxc:
TBabs(...) method of builtins.PyCapsule instance
TBabs(*args, **kwargs)
Overloaded function.
1. TBabs(pars: numpy.ndarray[numpy.float64], energies: numpy.ndarray[numpy.float64], spectrum: int = 1, initStr: str = '') -> numpy.ndarray[numpy.float64]
The XSPEC multiplicative TBabs model (1 parameter).
2. TBabs(pars: numpy.ndarray[numpy.float64], energies: numpy.ndarray[numpy.float64], out: numpy.ndarray[numpy.float64], spectrum: int = 1, initStr: str = '') -> numpy.ndarray[numpy.float64]
The XSPEC multiplicative TBabs model (1 parameter); inplace.
>>> x.TBabs([1], egrid)
tbvabs Version 2.3
Cosmic absorption with grains and H2, modified from
Wilms, Allen, & McCray, 2000, ApJ 542, 914-924
Questions: Joern Wilms
[email protected]
[email protected]
http://pulsar.sternwarte.uni-erlangen.de/wilms/research/tbabs/
PLEASE NOTICE:
To get the model described by the above paper
you will also have to set the abundances:
abund wilm
Note that this routine ignores the current cross section setting
as it always HAS to use the Verner cross sections as a baseline.
array([1.28407670e-175, 9.06810629e-062, 9.04157274e-029, 3.26034136e-016,
2.02047907e-010, 5.09045226e-007, 3.97658583e-005])
>>> x.TBabs([0.1], egrid)
array([3.24234405e-18, 7.86595867e-07, 1.56900525e-03, 2.82700325e-02,
1.07286588e-01, 2.34787860e-01, 3.63037122e-01])
>>> x.TBabs([0.01], egrid)
array([0.01782731, 0.24523089, 0.52427897, 0.70005576, 0.79993471,
0.86510249, 0.90363929])
Note that the return values have no units as this is an XSPEC multiplicative model.
The mkcflow additive model has a default of 0 for its redshift, but then
warns you about it!
mkcflow 5 0. 1.e20 C_xsmkcf add 0
lowT keV 0.1 0.0808 0.0808 79.9 79.9 0.001
highT keV 4. 0.0808 0.0808 79.9 79.9 0.001
Abundanc " " 1. 0. 0. 5. 5. 0.01
Redshift " " 0. -0.999 -0.999 10. 10. -0.01
$switch 1 0 0 1 1 -1
>>> x.mkcflow([0.1, 4, 1, 0, 1], np.arange(0.1, 0.8, 0.1))
XSVMCF: Require z > 0 for cooling flow models
array([0., 0., 0., 0., 0., 0.])
>>> x.mkcflow([0.1, 4, 1, 0, 1], np.arange(0.1, 0.8, 0.1))
XSVMCF: Require z > 0 for cooling flow models
array([0., 0., 0., 0., 0., 0.])
unless you set the chatter to 0:
>>> x.chatter(0)
>>> x.mkcflow([0.1, 4, 1, 0, 1], np.arange(0.1, 0.8, 0.1))
array([0., 0., 0., 0., 0., 0.])
>>>
The smaug model is an interesting one you have to set the XFLT keywords before using it. The model is
smaug 22 0.0E+00 1.0E+20 c_xsmaug add 0 1
kT.cc keV 1.0E+00 8.0E-02 1.0E-01 1.0E+01 1.0E+02 1.0E-02
kT.dt keV 1.0E+00 0.0E+00 0.0E+00 1.0E+01 1.0E+02 1.0E-02
kT.ix " " 0.0E+00 0.0E+00 0.0E+00 1.0E+01 1.0E+01 -1.0E-03
kT.ir Mpc 1.0E-01 1.0E-04 1.0E-04 1.0E+00 1.0E+00 -1.0E-03
kT.cx " " 5.0E-01 0.0E+00 0.0E+00 1.0E+01 1.0E+01 1.0E-03
kT.cr Mpc 1.0E-01 1.0E-04 1.0E-04 1.0E+01 2.0E+01 1.0E-02
kT.tx " " 0.0E+00 0.0E+00 0.0E+00 1.0E+01 1.0E+01 -1.0E-03
kT.tr Mpc 5.0E-01 1.0E-04 1.0E-04 1.0E+00 3.0E+00 -1.0E-02
nH.cc cm**-3 1.0E+00 1.0E-06 1.0E-06 3.0E+00 3.0E+00 -1.0E-02
nH.ff " " 1.0E-00 0.0E+00 0.0E+00 1.0E+00 1.0E+00 -1.0E-02
nH.cx " " 5.0E-01 0.0E+00 0.0E+00 1.0E+01 1.0E+01 1.0E-03
nH.cr Mpc 1.0E-01 1.0E-04 1.0E-04 1.0E+00 2.0E+00 1.0E-02
nH.gx " " 0.0E+00 0.0E+00 0.0E+00 1.0E+01 1.0E+01 -1.0E-03
nH.gr Mpc 2.0E-03 1.0E-04 1.0E-04 1.0E+01 2.0E+01 -1.0E-03
Ab.cc solar 1.0E+00 0.0E+00 0.0E+00 3.0E+00 5.0E+00 -1.0E-02
Ab.xx " " 0.0E+00 0.0E+00 0.0E+00 1.0E+01 1.0E+01 -1.0E-03
Ab.rr Mpc 1.0E-01 1.0E-04 1.0E-04 1.0E+00 1.0E+00 -1.0E-02
redshift " " 1.0E-02 1.0E-04 1.0E-04 1.0E+01 1.0E+01 -1.0E+00
meshpts " " 1.0E+01 1.0E+00 1.0E+00 1.0E+04 1.0E+04 -1.0E+00
rcutoff Mpc 2.0E+00 1.0E+00 1.0E+00 3.0E+00 3.0E+00 -1.0E-02
mode " " 1.0E+00 0.0E+00 0.0E+00 2.0E+00 2.0E+00 -1.0E+00
itype " " 2.0E+00 1.0E+00 1.0E+00 4.0E+00 4.0E+00 -1.0E+00
and when you try to run it the model fails (note that I use the
default parameter values apart for redshift, which is set to
something cosmologically interesting):
>>> pars = [0.4 if p.name == 'redshift' else p.default for p in x.info('smaug').parameters]
>>> x.smaug(pars, [0.1, 0.2, 0.3, 0.4, 0.5, 0.6])
***XSPEC Error: in function XSmaug: cannot find XFLTnnnn keyword for inner annulus for spectrum 1
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
RuntimeError: Caught an unknown exception!
We make probably make the error slightly nicer.
Now, we need to set
- cosmology setup
- the XFLT "inner", "outer", and "width" keywords
The cosmology is not set up by default:
>>> x.cosmology()
{'h0': 0.0, 'lambda0': 0.0, 'q0': 0.0}
So we change to:
>>> x.cosmology(h0=70, lambda0=0.73, q0=0)
and then set the XFLT keywords (making up these values as it depents
on the rcutoff parameter), but the result isn't quite what I
was expecting:
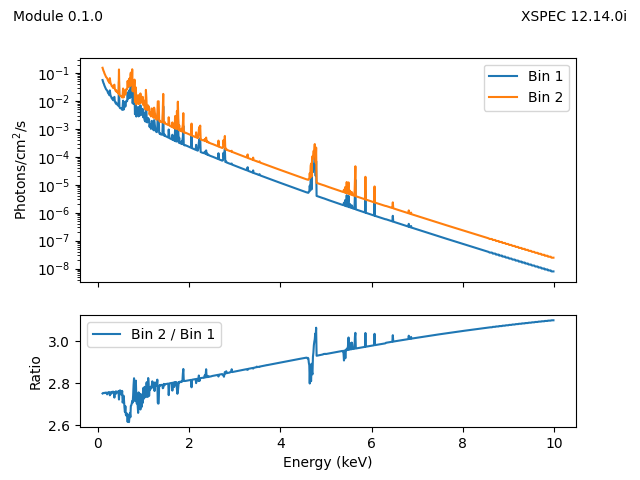
>>> x.setXFLT(1, {'inner': 0, 'outer': 0.1, 'width': 360})
>>> x.setXFLT(2, {'inner': 0.1, 'outer': 0.2, 'width': 360})
>>> egrid = np.arange(0.1, 7, 0.01)
>>> y1 = x.smaug(energies=egrid, pars=pars, spectrum=1)
>>> y2 = x.smaug(energies=egrid, pars=pars, spectrum=2)
We can plot this:
>>> emid = (egrid[:-1] + egrid[1:]) / 2
>>> plt.plot(emid, y1, label='Bin 1')
>>> plt.plot(emid, y2, label='Bin 2')
>>> plt.yscale('log')
>>> plt.legend()
>>> plt.xlabel('Energy (keV)')
>>> plt.ylabel('Photons/cm$^2$/s')
to create (although this is a slightly-more-complex version):
The cflux convolution model changes the normalization of the input
model so it matches 10^lg10Flux for the Emin to Emax range.
cflux 3 0. 1.e20 C_cflux con 0
Emin "keV" 0.5 0.0 0.0 1e6 1e6 -0.1
Emax "keV" 10.0 0.0 0.0 1e6 1e6 -0.1
lg10Flux "cgs" -12.0 -100.0 -100.0 100. 100. 0.01
Let's try to convolve a powerlaw over the range 0.5 to 10 keV:
>>> help(x.cflux)
Help on built-in function cflux in module xspec_models_cxc:
cflux(...) method of builtins.PyCapsule instance
cflux(pars: numpy.ndarray[numpy.float64], energies: numpy.ndarray[numpy.float64], model: numpy.ndarray[numpy.float64], spectrum: int = 1, initStr: str = '') -> numpy.ndarray[numpy.float64]
The XSPEC convolution cflux model (3 parameters); inplace.
>>> egrid = np.arange(0.4, 10.2, 0.1)
>>> pars = [0.5, 10, -12]
>>> y1 = x.powerlaw(pars=[-1.7], energies=egrid)
>>> y2 = x.cflux(pars=pars, energies=egrid, model=y1.copy())
Note that convolution models always over-write the model
argument - so if we had used model=y1 rather than model=y1.copy()
then y1 would have been changed (which is normally okay, but in this
example I wanted to compare the input and output arrays).
There is a subtly to using the model argument: it must have the same
date type as the convolution model expects - which can be found by
checking help(x.<convolution model>) or x.info(<convolution model)>.language - otherwise it will not be updated. In this case
cflux uses float64 so it would work out:
>>> egrid.dtype
dtype('float64')
>>> y1.dtype
dtype('float64')
>>> y2.dtype
dtype('float64')
>>> x.info('cflux').language
<LanguageStyle.CppStyle8: 1>
Now, we need to sum up y2 over the range 0.5 to 10 keV,
which thanks to the grid I chose, is all-but the first and
last bins:
>>> egrid[:3], egrid[-3:]
(array([0.4, 0.5, 0.6]), array([ 9.9, 10. , 10.1]))
We shall use the mid-point of each bin for converting from photons/cm^2/s to erg/cm^2/s, and as I can never remember the conversion factor, let's calculate it
>>> emid_kev = (egrid[1:-2] + egrid[2:-1]) / 2
>>> import astropy.units as u
>>> ((1 * u.keV) / (1 * u.erg)).decompose()
<Quantity 1.60217663e-09>
>>> conv = ((1 * u.keV) / (1 * u.erg)).decompose().value
>>> emid_kev = (egrid[1:-2] + egrid[2:-1]) / 2
With this we can compare the flux of the model before and after
convolution by cflux. We can see the result is 1e-12 which matches
the lg10Flux parameter:
>>> (y1[1:-1] * emid_kev).sum() * conv
2.170144702398361e-06
>>> (y2[1:-1] * emid_kev).sum() * conv
9.999904093891366e-13
The interface is somewhat "janky" - to use an official term - and relies on the user understanding the model metadata. This will be improved upon, but for now you need to know
- what type it is ("add", "mul", or "exp" for "atable", "mtable", or "etable")
- what the parameters are (includng whether redshift is added)
- what the parameter ranges are
With that, the RCS.mod table model - an additive model with three parameters with default values of [1, 0.1, 0.1] and no redshift parameter - can be evaluated:
>>> infile = 'RCS.mod'
>>> mtype = 'add'
>>> pars = [1, 0.1, 0.1]
>>> y = x.tableModel(table=infile, table_type=mtype, pars=pars, energies=egrid)
It is very easy to crash the interpreter - for example
- if you send in too few parameters
- if you send in a parameter outside the hard range for that parameter
>>> help(x.tableModel)
Help on built-in function tableModel in module xspec_models_cxc._compiled:
tableModel(...) method of builtins.PyCapsule instance
tableModel(*args, **kwargs)
Overloaded function.
1. tableModel(table: str, table_type: str, pars: numpy.ndarray[numpy.float32], energies: numpy.ndarray[numpy.float32], spectrum: int = 1) -> numpy.ndarray[numpy.float32]
XSPEC table model.
2. tableModel(table: str, table_type: str, pars: numpy.ndarray[numpy.float32], energies: numpy.ndarray[numpy.float32], model: numpy.ndarray[numpy.float32], spectrum: int = 1) -> numpy.ndarray[numpy.float32]
XSPEC table model; inplace.
Version 0.0.24 and later allows us to "directly" use the C++ interface, at the expense of using the x.RealArray type rather than NumPy arrays. The following models
>>> x.list_models(language=x.LanguageStyle.CppStyle8)
['TBabs', 'TBfeo', 'TBgas', 'TBgrain', 'TBpcf', 'TBrel', 'TBvarabs',
'absori', 'acisabs', 'agauss', 'apec', 'bapec', 'bexrav', 'bexriv',
...
'xscat', 'zTBabs', 'zagauss', 'zashift', 'zbknpower', 'zcutoffpl',
'zgauss', 'zkerrbb', 'zlogpar', 'zmshift', 'zpowerlw', 'zxipcf']
all have a "_" variant such as
>>> help(x.TBabs_)
Help on built-in function TBabs_ in module xspec_models_cxc._compiled:
TBabs_(...) method of builtins.PyCapsule instance
TBabs_(pars: xspec_models_cxc._compiled.RealArray, energies: xspec_models_cxc._compiled.RealArray, out: xspec_models_cxc._compiled.RealArray, spectrum: int = 1, initStr: str = '') -> xspec_models_cxc._compiled.RealArray
The XSPEC multiplicative TBabs model (1 parameter); RealArray, inplace.
which use this interface. The x.RealArray type
>>> help(x.RealArray)
Help on class RealArray in module xspec_models_cxc._compiled:
class RealArray(pybind11_builtins.pybind11_object)
| Method resolution order:
| RealArray
| pybind11_builtins.pybind11_object
| builtins.object
|
| Methods defined here:
|
| __getitem__(...)
| __getitem__(self: xspec_models_cxc._compiled.RealArray, arg0: int) -> float
|
| __init__(...)
| __init__(*args, **kwargs)
| Overloaded function.
|
| 1. __init__(self: xspec_models_cxc._compiled.RealArray, arg0: int) -> None
|
| Create an array of n zeros.
|
| 2. __init__(self: xspec_models_cxc._compiled.RealArray, arg0: numpy.ndarray[numpy.float64]) -> None
|
| Copy the data into an array.
|
...
will convert a one-dimensional sequence into something that can be sent to these routines (or, when given a non-negative integer, willreturn an array of 0's of that length):
>>> pars = x.RealArray([p.default for p in x.info('TBabs').parameters])
>>> egrid = x.RealArray(np.arange(0.1, 1, 0.1))
>>> out = x.RealArray(len(egrid) - 1)
>>> y = x.TBabs_(energies=egrid, pars=pars, out=out)
tbvabs Version 2.3
Cosmic absorption with grains and H2, modified from
Wilms, Allen, & McCray, 2000, ApJ 542, 914-924
Questions: Joern Wilms
[email protected]
[email protected]
http://pulsar.sternwarte.uni-erlangen.de/wilms/research/tbabs/
PLEASE NOTICE:
To get the model described by the above paper
you will also have to set the abundances:
abund wilm
Note that this routine ignores the current cross section setting
as it always HAS to use the Verner cross sections as a baseline.
>>> y is out
True
>>> y
[2.3158e-154, 2.41183e-26, 5.47728e-10, 4.24854e-05, 0.000648813, 0.00195077, 0.0112892, 0.0261219]
Note that the out argument is changed, as well as being returned
(in the same way it works for the inplace versions of this).
This version could have been overloaded onto the model name but I
wanted to make it easier to see if things like the symbol overloading
make any significant difference to the runtime of a model. Switching
to IPython to take advantage of %timeit:
In [1]: import numpy as np
In [2]: import xspec_models_cxc as x
In [3]: pars1 = [p.default for p in x.info('bvapec').parameters]
In [4]: pars2 = x.RealArray(pars1)
In [5]: egrid1 = np.arange(0.1, 11, 0.001)
In [6]: egrid2 = x.RealArray(egrid1)
In [7]: y1 = x.bvapec(energies=egrid1, pars=pars1)
Reading APEC data from 3.0.9
In [8]: out2 = x.RealArray(len(egrid2) - 1)
In [9]: y2 = x.bvapec_(energies=egrid2, pars=pars2, out=out2)
In [10]: (y1 == y2).all()
Out[10]: True
In [11]: %timeit y1 = x.bvapec(energies=egrid1, pars=pars1)
6.85 ms ± 39.9 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
In [12]: %timeit y2 = x.bvapec_(energies=egrid2, pars=pars2, out=out2)
7.13 ms ± 26.3 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
So, for this simple case it doesn't seem worth using the RealArray
version (but perhaps not returning the array and other changes could
alter this).
One question is how much time is spent in marshallnig data to and from the compiled code? One point of view is "the ability to evaluate a model from Python is worth the cost", but I'd like to see what we can do to mitigate this cost. There are various avenues worth persuing, and this module does some of the following:
-
handle the model evaluation completely in C++ (that is, we encode an AST for expressions like
"phabs * (powerlaw + apec + gaussian + gaussian)"
and evalaute the AST in C++).
I'm interested in this, but haven't got around to working on it yet, and there are issues in how we send in the parameters: see #12
-
the current Sherpa-inspired interface creates the output array each time we call the model, leading to a lot of memory churn. Perhaps we can re-use arrays rather than re-creating them.
Many of the models have an "inplace" variant which has the out argument which supports this. At the moment I haven't seen any evidence of this making model evaluation faster, but perhaps it needs to be run through a typical "fit" where we may see benefits from reduced memory overhead.
Notes:
- The convolution models only support the inplace varsion.
- Use of the out argument is easy to mess up - see #14
-
the different XSPEC models have different "preferred" interfaces (FORTRAN, C, or C++) and using the correct one should reduce the run-time of the model.
Initial tests suggest that the conversion cost is not a significant part of the run-time cost of a model, but I can't guarantee it.
Note that we currently bind to the correct versions for FORTRAN (single precision) and C models, but the C++ models (which, in XSPEC 12.14.1 is 197 of the 293 models we support) are handled using the C interface. To see if this is a problem I have added (in version 0.0.24) the RealArray interface discussed above, but it doesn't seem to make much difference, although this claim needs to be tested.