+
+
+
+
+02:00
+ +
+::::::::::::::::::::::::::::::::::::: challenge
+
+### Challenge 1: Import line and point vector layers
+
+Read in `delft-streets.shp` and `delft-leisure.shp` and assign them to `lines_Delft` and `point_Delft` respectively. Answer the following questions:
+
+1. What type of R spatial object is created when you import each layer?
+2. What is the CRS and extent for each object?
+3. Do the files contain points, lines, or polygons?
+4. How many features are in each file?
+
+:::::::::::::::::::::::: solution
+
+
+```r
+lines_Delft <- st_read("data/delft-streets.shp")
+point_Delft <- st_read("data/delft-leisure.shp")
+```
+
+We can check the type of data with the `class()` function from base R. Both `lines_Delft` and `point_Delft` are objects of class `"sf"`, which extends the `"data.frame"` class.
+
+
+```r
+class(lines_Delft)
+```
+
+```{.output}
+[1] "sf" "data.frame"
+```
+
+```r
+class(point_Delft)
+```
+
+```{.output}
+[1] "sf" "data.frame"
+```
+
+`lines_Delft` and `point_Delft` are in the correct CRS.
+
+
+```r
+st_crs(lines_Delft)$epsg
+```
+
+```{.output}
+[1] 28992
+```
+
+```r
+st_crs(point_Delft)$epsg
+```
+
+```{.output}
+[1] 28992
+```
+
+When looking at the bounding boxes with the `st_bbox()` function, we see the spatial extent of the two objects in a projected CRS using meters as units. `lines_Delft()` and `point_Delft` have similar extents.
+
+
+```r
+st_bbox(lines_Delft)
+```
+
+```{.output}
+ xmin ymin xmax ymax
+ 81759.58 441223.13 89081.41 449845.81
+```
+
+```r
+st_bbox(point_Delft)
+```
+
+```{.output}
+ xmin ymin xmax ymax
+ 81863.21 442621.15 87370.15 449345.08
+```
+
+:::::::::::::::::::::::::::::::::
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+
+
+::::::::::::::::::::::::::::::::::::: keypoints
+
+- Metadata for vector layers include geometry type, CRS, and extent.
+- Load spatial objects into R with the `st_read()` function.
+- Spatial objects can be plotted directly with `ggplot` using the `geom_sf()` function. No need to convert to a data frame.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
diff --git a/10-explore-and-plot-by-vector-layer-attributes.md b/10-explore-and-plot-by-vector-layer-attributes.md
new file mode 100644
index 00000000..d65a938d
--- /dev/null
+++ b/10-explore-and-plot-by-vector-layer-attributes.md
@@ -0,0 +1,803 @@
+---
+title: 'Explore and plot by vector layer attributes'
+teaching: 20
+exercises: 10
+---
+
+
+
+
+:::::::::::::::::::::::::::::::::::::: questions
+
+- How can I examine the attributes of a vector layer?
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::::::::::::::::::::::::::::::::::::: objectives
+
+- Query attributes of a vector object.
+
+- Subset vector objects using specific attribute values.
+
+- Plot a vector feature, colored by unique attribute values.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+# Query Vector Feature Metadata
+
+Let's have a look at the content of the loaded data, starting with `lines_Delft`. In essence, an `"sf"` object is a data.frame with a "sticky" geometry column and some extra metadata, like the CRS, extent and geometry type we examined earlier.
+
+
+```r
+lines_Delft
+```
+
+```{.output}
+Simple feature collection with 11244 features and 2 fields
+Geometry type: LINESTRING
+Dimension: XY
+Bounding box: xmin: 81759.58 ymin: 441223.1 xmax: 89081.41 ymax: 449845.8
+Projected CRS: Amersfoort / RD New
+First 10 features:
+ osm_id highway geometry
+1 4239535 cycleway LINESTRING (86399.68 448599...
+2 4239536 cycleway LINESTRING (85493.66 448740...
+3 4239537 cycleway LINESTRING (85493.66 448740...
+4 4239620 footway LINESTRING (86299.01 448536...
+5 4239621 footway LINESTRING (86307.35 448738...
+6 4239674 footway LINESTRING (86299.01 448536...
+7 4310407 service LINESTRING (84049.47 447778...
+8 4310808 steps LINESTRING (84588.83 447828...
+9 4348553 footway LINESTRING (84527.26 447861...
+10 4348575 footway LINESTRING (84500.15 447255...
+```
+
+This means that we can examine and manipulate them as data frames. For instance, we can look at the number of variables (columns in a data frame) with `ncol()`.
+
+
+```r
+ncol(lines_Delft)
+```
+
+```{.output}
+[1] 3
+```
+
+In the case of `point_Delft` those columns are `"osm_id"`, `"highway"` and `"geometry"`. We can check the names of the columns with the base R function `names()`.
+
+
+```r
+names(lines_Delft)
+```
+
+```{.output}
+[1] "osm_id" "highway" "geometry"
+```
+
+::: callout
+
+Note that in R the geometry is just another column and counts towards the number
+returned by `ncol()`. This is different from GIS software with graphical user
+interfaces, where the geometry is displayed in a viewport not as a column in the
+attribute table.
+
+:::
+
+We can also preview the content of the object by looking at the first 6 rows with the `head()` function, which is in the case of an `sf` object the same as examining the object directly.
+
+
+```r
+head(lines_Delft)
+```
+
+```{.output}
+Simple feature collection with 6 features and 2 fields
+Geometry type: LINESTRING
+Dimension: XY
+Bounding box: xmin: 85107.1 ymin: 448400.3 xmax: 86399.68 ymax: 449076.2
+Projected CRS: Amersfoort / RD New
+ osm_id highway geometry
+1 4239535 cycleway LINESTRING (86399.68 448599...
+2 4239536 cycleway LINESTRING (85493.66 448740...
+3 4239537 cycleway LINESTRING (85493.66 448740...
+4 4239620 footway LINESTRING (86299.01 448536...
+5 4239621 footway LINESTRING (86307.35 448738...
+6 4239674 footway LINESTRING (86299.01 448536...
+```
+
+
+## Explore values within one attribute
+
+Using the `$` operator, we can examine the content of a single field of our lines feature.
+
+We can see the contents of the `highway` field of our lines feature:
+
+
+```r
+head(lines_Delft$highway, 10)
+```
+
+```{.output}
+ [1] "cycleway" "cycleway" "cycleway" "footway" "footway" "footway"
+ [7] "service" "steps" "footway" "footway"
+```
+
+To see only unique values within the `highway` field, we can use the `unique()` function. This function extracts all possible values of a character variable.
+
+
+```r
+unique(lines_Delft$highway)
+```
+
+```{.output}
+ [1] "cycleway" "footway" "service" "steps"
+ [5] "residential" "unclassified" "construction" "secondary"
+ [9] "busway" "living_street" "motorway_link" "tertiary"
+[13] "track" "motorway" "path" "pedestrian"
+[17] "primary" "bridleway" "trunk" "tertiary_link"
+[21] "services" "secondary_link" "trunk_link" "primary_link"
+[25] "platform" "proposed" NA
+```
+
+:::::::::::::::::::::::: callout
+
+R is also able to handle categorical variables called factors. With factors, we can use the `levels()` function to show unique values. To examine unique values of the `highway` variable this way, we have to first transform it into a factor with the `factor()` function:
+
+
+```r
+levels(factor(lines_Delft$highway))
+```
+
+```{.output}
+ [1] "bridleway" "busway" "construction" "cycleway"
+ [5] "footway" "living_street" "motorway" "motorway_link"
+ [9] "path" "pedestrian" "platform" "primary"
+[13] "primary_link" "proposed" "residential" "secondary"
+[17] "secondary_link" "service" "services" "steps"
+[21] "tertiary" "tertiary_link" "track" "trunk"
+[25] "trunk_link" "unclassified"
+```
+
+::::::::::::::::::::::::::::::::
+
+::::::::::::::::::::::::::::::::::::: challenge
+
+## Challenge: Attributes for different spatial classes
+
+
+Explore the attributes associated with the `point_Delft` spatial object.
+
+1. How many attributes does it have?
+2. What types of leisure points do the points represent? Give three examples.
+3. Which of the following is NOT an attribute of the `point_Delft` data object?
+
+ A) location B) leisure C) osm_id
+
+:::::::::::::::::::::::: solution
+
+1. To find the number of attributes, we use the `ncol()` function:
+
+
+```r
+ncol(point_Delft)
+```
+
+```{.output}
+[1] 3
+```
+
+2. The types of leisure point are in the column named `leisure`.
+
+Using the `head()` function which displays 6 rows by default, we only see two values and `NA`s.
+
+
+```r
+head(point_Delft)
+```
+
+```{.output}
+Simple feature collection with 6 features and 2 fields
+Geometry type: POINT
+Dimension: XY
+Bounding box: xmin: 83839.59 ymin: 443827.4 xmax: 84967.67 ymax: 447475.5
+Projected CRS: Amersfoort / RD New
+ osm_id leisure geometry
+1 472312297 picnic_table POINT (84144.72 443827.4)
+2 480470725 marina POINT (84967.67 446120.1)
+3 484697679
+
+::::::::::::::::::::::::::::::::::::: challenge
+
+### Challenge 1: Import line and point vector layers
+
+Read in `delft-streets.shp` and `delft-leisure.shp` and assign them to `lines_Delft` and `point_Delft` respectively. Answer the following questions:
+
+1. What type of R spatial object is created when you import each layer?
+2. What is the CRS and extent for each object?
+3. Do the files contain points, lines, or polygons?
+4. How many features are in each file?
+
+:::::::::::::::::::::::: solution
+
+
+```r
+lines_Delft <- st_read("data/delft-streets.shp")
+point_Delft <- st_read("data/delft-leisure.shp")
+```
+
+We can check the type of data with the `class()` function from base R. Both `lines_Delft` and `point_Delft` are objects of class `"sf"`, which extends the `"data.frame"` class.
+
+
+```r
+class(lines_Delft)
+```
+
+```{.output}
+[1] "sf" "data.frame"
+```
+
+```r
+class(point_Delft)
+```
+
+```{.output}
+[1] "sf" "data.frame"
+```
+
+`lines_Delft` and `point_Delft` are in the correct CRS.
+
+
+```r
+st_crs(lines_Delft)$epsg
+```
+
+```{.output}
+[1] 28992
+```
+
+```r
+st_crs(point_Delft)$epsg
+```
+
+```{.output}
+[1] 28992
+```
+
+When looking at the bounding boxes with the `st_bbox()` function, we see the spatial extent of the two objects in a projected CRS using meters as units. `lines_Delft()` and `point_Delft` have similar extents.
+
+
+```r
+st_bbox(lines_Delft)
+```
+
+```{.output}
+ xmin ymin xmax ymax
+ 81759.58 441223.13 89081.41 449845.81
+```
+
+```r
+st_bbox(point_Delft)
+```
+
+```{.output}
+ xmin ymin xmax ymax
+ 81863.21 442621.15 87370.15 449345.08
+```
+
+:::::::::::::::::::::::::::::::::
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+
+
+::::::::::::::::::::::::::::::::::::: keypoints
+
+- Metadata for vector layers include geometry type, CRS, and extent.
+- Load spatial objects into R with the `st_read()` function.
+- Spatial objects can be plotted directly with `ggplot` using the `geom_sf()` function. No need to convert to a data frame.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
diff --git a/10-explore-and-plot-by-vector-layer-attributes.md b/10-explore-and-plot-by-vector-layer-attributes.md
new file mode 100644
index 00000000..d65a938d
--- /dev/null
+++ b/10-explore-and-plot-by-vector-layer-attributes.md
@@ -0,0 +1,803 @@
+---
+title: 'Explore and plot by vector layer attributes'
+teaching: 20
+exercises: 10
+---
+
+
+
+
+:::::::::::::::::::::::::::::::::::::: questions
+
+- How can I examine the attributes of a vector layer?
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::::::::::::::::::::::::::::::::::::: objectives
+
+- Query attributes of a vector object.
+
+- Subset vector objects using specific attribute values.
+
+- Plot a vector feature, colored by unique attribute values.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+# Query Vector Feature Metadata
+
+Let's have a look at the content of the loaded data, starting with `lines_Delft`. In essence, an `"sf"` object is a data.frame with a "sticky" geometry column and some extra metadata, like the CRS, extent and geometry type we examined earlier.
+
+
+```r
+lines_Delft
+```
+
+```{.output}
+Simple feature collection with 11244 features and 2 fields
+Geometry type: LINESTRING
+Dimension: XY
+Bounding box: xmin: 81759.58 ymin: 441223.1 xmax: 89081.41 ymax: 449845.8
+Projected CRS: Amersfoort / RD New
+First 10 features:
+ osm_id highway geometry
+1 4239535 cycleway LINESTRING (86399.68 448599...
+2 4239536 cycleway LINESTRING (85493.66 448740...
+3 4239537 cycleway LINESTRING (85493.66 448740...
+4 4239620 footway LINESTRING (86299.01 448536...
+5 4239621 footway LINESTRING (86307.35 448738...
+6 4239674 footway LINESTRING (86299.01 448536...
+7 4310407 service LINESTRING (84049.47 447778...
+8 4310808 steps LINESTRING (84588.83 447828...
+9 4348553 footway LINESTRING (84527.26 447861...
+10 4348575 footway LINESTRING (84500.15 447255...
+```
+
+This means that we can examine and manipulate them as data frames. For instance, we can look at the number of variables (columns in a data frame) with `ncol()`.
+
+
+```r
+ncol(lines_Delft)
+```
+
+```{.output}
+[1] 3
+```
+
+In the case of `point_Delft` those columns are `"osm_id"`, `"highway"` and `"geometry"`. We can check the names of the columns with the base R function `names()`.
+
+
+```r
+names(lines_Delft)
+```
+
+```{.output}
+[1] "osm_id" "highway" "geometry"
+```
+
+::: callout
+
+Note that in R the geometry is just another column and counts towards the number
+returned by `ncol()`. This is different from GIS software with graphical user
+interfaces, where the geometry is displayed in a viewport not as a column in the
+attribute table.
+
+:::
+
+We can also preview the content of the object by looking at the first 6 rows with the `head()` function, which is in the case of an `sf` object the same as examining the object directly.
+
+
+```r
+head(lines_Delft)
+```
+
+```{.output}
+Simple feature collection with 6 features and 2 fields
+Geometry type: LINESTRING
+Dimension: XY
+Bounding box: xmin: 85107.1 ymin: 448400.3 xmax: 86399.68 ymax: 449076.2
+Projected CRS: Amersfoort / RD New
+ osm_id highway geometry
+1 4239535 cycleway LINESTRING (86399.68 448599...
+2 4239536 cycleway LINESTRING (85493.66 448740...
+3 4239537 cycleway LINESTRING (85493.66 448740...
+4 4239620 footway LINESTRING (86299.01 448536...
+5 4239621 footway LINESTRING (86307.35 448738...
+6 4239674 footway LINESTRING (86299.01 448536...
+```
+
+
+## Explore values within one attribute
+
+Using the `$` operator, we can examine the content of a single field of our lines feature.
+
+We can see the contents of the `highway` field of our lines feature:
+
+
+```r
+head(lines_Delft$highway, 10)
+```
+
+```{.output}
+ [1] "cycleway" "cycleway" "cycleway" "footway" "footway" "footway"
+ [7] "service" "steps" "footway" "footway"
+```
+
+To see only unique values within the `highway` field, we can use the `unique()` function. This function extracts all possible values of a character variable.
+
+
+```r
+unique(lines_Delft$highway)
+```
+
+```{.output}
+ [1] "cycleway" "footway" "service" "steps"
+ [5] "residential" "unclassified" "construction" "secondary"
+ [9] "busway" "living_street" "motorway_link" "tertiary"
+[13] "track" "motorway" "path" "pedestrian"
+[17] "primary" "bridleway" "trunk" "tertiary_link"
+[21] "services" "secondary_link" "trunk_link" "primary_link"
+[25] "platform" "proposed" NA
+```
+
+:::::::::::::::::::::::: callout
+
+R is also able to handle categorical variables called factors. With factors, we can use the `levels()` function to show unique values. To examine unique values of the `highway` variable this way, we have to first transform it into a factor with the `factor()` function:
+
+
+```r
+levels(factor(lines_Delft$highway))
+```
+
+```{.output}
+ [1] "bridleway" "busway" "construction" "cycleway"
+ [5] "footway" "living_street" "motorway" "motorway_link"
+ [9] "path" "pedestrian" "platform" "primary"
+[13] "primary_link" "proposed" "residential" "secondary"
+[17] "secondary_link" "service" "services" "steps"
+[21] "tertiary" "tertiary_link" "track" "trunk"
+[25] "trunk_link" "unclassified"
+```
+
+::::::::::::::::::::::::::::::::
+
+::::::::::::::::::::::::::::::::::::: challenge
+
+## Challenge: Attributes for different spatial classes
+
+
+Explore the attributes associated with the `point_Delft` spatial object.
+
+1. How many attributes does it have?
+2. What types of leisure points do the points represent? Give three examples.
+3. Which of the following is NOT an attribute of the `point_Delft` data object?
+
+ A) location B) leisure C) osm_id
+
+:::::::::::::::::::::::: solution
+
+1. To find the number of attributes, we use the `ncol()` function:
+
+
+```r
+ncol(point_Delft)
+```
+
+```{.output}
+[1] 3
+```
+
+2. The types of leisure point are in the column named `leisure`.
+
+Using the `head()` function which displays 6 rows by default, we only see two values and `NA`s.
+
+
+```r
+head(point_Delft)
+```
+
+```{.output}
+Simple feature collection with 6 features and 2 fields
+Geometry type: POINT
+Dimension: XY
+Bounding box: xmin: 83839.59 ymin: 443827.4 xmax: 84967.67 ymax: 447475.5
+Projected CRS: Amersfoort / RD New
+ osm_id leisure geometry
+1 472312297 picnic_table POINT (84144.72 443827.4)
+2 480470725 marina POINT (84967.67 446120.1)
+3 484697679  +
+Map of cycleways in Delft.
+ +
+5. Same steps with pedestrian streets
+
+
+```r
+pedestrian_Delft <- lines_Delft %>%
+ filter(highway == "pedestrian")
+
+pedestrian_Delft %>%
+ mutate(length = st_length(.)) %>%
+ select(everything(), geometry) %>%
+ summarise(total_length = sum(length))
+```
+
+```{.output}
+Simple feature collection with 1 feature and 1 field
+Geometry type: MULTILINESTRING
+Dimension: XY
+Bounding box: xmin: 82388.15 ymin: 444400.2 xmax: 85875.95 ymax: 447987.8
+Projected CRS: Amersfoort / RD New
+ total_length geometry
+1 12778.84 [m] MULTILINESTRING ((85538.03 ...
+```
+
+```r
+nrow(pedestrian_Delft)
+```
+
+```{.output}
+[1] 234
+```
+
+
+```r
+ggplot() +
+ geom_sf(data = pedestrian_Delft) +
+ labs(title = "Slow mobility network", subtitle = "Pedestrian") +
+ coord_sf(datum = st_crs(28992))
+```
+
+
+
+5. Same steps with pedestrian streets
+
+
+```r
+pedestrian_Delft <- lines_Delft %>%
+ filter(highway == "pedestrian")
+
+pedestrian_Delft %>%
+ mutate(length = st_length(.)) %>%
+ select(everything(), geometry) %>%
+ summarise(total_length = sum(length))
+```
+
+```{.output}
+Simple feature collection with 1 feature and 1 field
+Geometry type: MULTILINESTRING
+Dimension: XY
+Bounding box: xmin: 82388.15 ymin: 444400.2 xmax: 85875.95 ymax: 447987.8
+Projected CRS: Amersfoort / RD New
+ total_length geometry
+1 12778.84 [m] MULTILINESTRING ((85538.03 ...
+```
+
+```r
+nrow(pedestrian_Delft)
+```
+
+```{.output}
+[1] 234
+```
+
+
+```r
+ggplot() +
+ geom_sf(data = pedestrian_Delft) +
+ labs(title = "Slow mobility network", subtitle = "Pedestrian") +
+ coord_sf(datum = st_crs(28992))
+```
+
+ +
+:::
+
+:::
+
+## Customize plots
+
+Let's say that we want to color different road types with different colors and that we want to determine those colors.
+
+
+```r
+unique(lines_Delft$highway)
+```
+
+```{.output}
+ [1] "cycleway" "footway" "service" "steps"
+ [5] "residential" "unclassified" "construction" "secondary"
+ [9] "busway" "living_street" "motorway_link" "tertiary"
+[13] "track" "motorway" "path" "pedestrian"
+[17] "primary" "bridleway" "trunk" "tertiary_link"
+[21] "services" "secondary_link" "trunk_link" "primary_link"
+[25] "platform" "proposed" NA
+```
+
+If we look at all the unique values of the highway field of our street network we see more than 20 values. Let's focus on a subset of four values to illustrate the use of distinct colors. We use a piped expression in which we only filter the rows of our data frame that have one of the four given values `"motorway"`, `"primary"`, `"secondary"`, and `"cycleway"`. We also make sure that the highway column is a factor column.
+
+
+```r
+road_types <- c("motorway", "primary", "secondary", "cycleway")
+
+lines_Delft_selection <- lines_Delft %>%
+ filter(highway %in% road_types) %>%
+ mutate(highway = factor(highway, levels = road_types))
+```
+
+Next we define the four colors we want to use, one for each type of road in our vector object. Note that in R you can use named colors like `"blue"`, `"green"`, `"navy"`, and `"purple"`. A full list of named colors can be listed with the `colors()` function.
+
+
+```r
+road_colors <- c("blue", "green", "navy", "purple")
+```
+
+We can use the defined color palette in ggplot.
+
+
+```r
+ggplot(data = lines_Delft_selection) +
+ geom_sf(aes(color = highway)) +
+ scale_color_manual(values = road_colors) +
+ labs(color = 'Road Type',
+ title = "Road network of Delft",
+ subtitle = "Roads & Cycleways") +
+ coord_sf(datum = st_crs(28992))
+```
+
+
+
+:::
+
+:::
+
+## Customize plots
+
+Let's say that we want to color different road types with different colors and that we want to determine those colors.
+
+
+```r
+unique(lines_Delft$highway)
+```
+
+```{.output}
+ [1] "cycleway" "footway" "service" "steps"
+ [5] "residential" "unclassified" "construction" "secondary"
+ [9] "busway" "living_street" "motorway_link" "tertiary"
+[13] "track" "motorway" "path" "pedestrian"
+[17] "primary" "bridleway" "trunk" "tertiary_link"
+[21] "services" "secondary_link" "trunk_link" "primary_link"
+[25] "platform" "proposed" NA
+```
+
+If we look at all the unique values of the highway field of our street network we see more than 20 values. Let's focus on a subset of four values to illustrate the use of distinct colors. We use a piped expression in which we only filter the rows of our data frame that have one of the four given values `"motorway"`, `"primary"`, `"secondary"`, and `"cycleway"`. We also make sure that the highway column is a factor column.
+
+
+```r
+road_types <- c("motorway", "primary", "secondary", "cycleway")
+
+lines_Delft_selection <- lines_Delft %>%
+ filter(highway %in% road_types) %>%
+ mutate(highway = factor(highway, levels = road_types))
+```
+
+Next we define the four colors we want to use, one for each type of road in our vector object. Note that in R you can use named colors like `"blue"`, `"green"`, `"navy"`, and `"purple"`. A full list of named colors can be listed with the `colors()` function.
+
+
+```r
+road_colors <- c("blue", "green", "navy", "purple")
+```
+
+We can use the defined color palette in ggplot.
+
+
+```r
+ggplot(data = lines_Delft_selection) +
+ geom_sf(aes(color = highway)) +
+ scale_color_manual(values = road_colors) +
+ labs(color = 'Road Type',
+ title = "Road network of Delft",
+ subtitle = "Roads & Cycleways") +
+ coord_sf(datum = st_crs(28992))
+```
+
+ +
+## Adjust line width
+
+Earlier we adjusted the line width universally. We can also adjust line widths for every factor level. Note that in this case the `size` argument, like the `color` argument, are within the `aes()` mapping function. This means that the values of that visual property will be mapped from a variable of the object that is being plotted.
+
+
+```r
+line_widths <- c(1, 0.75, 0.5, 0.25)
+```
+
+
+```r
+ggplot(data = lines_Delft_selection) +
+ geom_sf(aes(color = highway, linewidth = highway)) +
+ scale_color_manual(values = road_colors) +
+ labs(color = 'Road Type',
+ linewidth = 'Road Type',
+ title = "Mobility network of Delft",
+ subtitle = "Roads & Cycleways") +
+ scale_linewidth_manual(values = line_widths) +
+ coord_sf(datum = st_crs(28992))
+```
+
+
+
+## Adjust line width
+
+Earlier we adjusted the line width universally. We can also adjust line widths for every factor level. Note that in this case the `size` argument, like the `color` argument, are within the `aes()` mapping function. This means that the values of that visual property will be mapped from a variable of the object that is being plotted.
+
+
+```r
+line_widths <- c(1, 0.75, 0.5, 0.25)
+```
+
+
+```r
+ggplot(data = lines_Delft_selection) +
+ geom_sf(aes(color = highway, linewidth = highway)) +
+ scale_color_manual(values = road_colors) +
+ labs(color = 'Road Type',
+ linewidth = 'Road Type',
+ title = "Mobility network of Delft",
+ subtitle = "Roads & Cycleways") +
+ scale_linewidth_manual(values = line_widths) +
+ coord_sf(datum = st_crs(28992))
+```
+
+ +
+::: challenge
+
+# Plot line width by attribute
+
+In the example above, we set the line widths to be 1, 0.75, 0.5, and 0.25. In our case line thicknesses are consistent with the hierarchy of the selected road types, but in some cases we might want to show a different hierarchy.
+
+Let’s create another plot where we show the different line types with the following thicknesses:
+
+- motorways linewidth = 0.25
+- primary linewidth = 0.75
+- secondary linewidth = 0.5
+- cycleway linewidth = 1
+
+::: solution
+
+
+```r
+levels(factor(lines_Delft_selection$highway))
+```
+
+```{.output}
+[1] "motorway" "primary" "secondary" "cycleway"
+```
+
+
+```r
+line_width <- c(0.25, 0.75, 0.5, 1)
+```
+
+
+```r
+ggplot(data = lines_Delft_selection) +
+ geom_sf(aes(linewidth = highway)) +
+ scale_linewidth_manual(values = line_width) +
+ labs(title = "Mobility network of Delft",
+ subtitle = "Roads & Cycleways - Line width varies") +
+ coord_sf(datum = st_crs(28992))
+```
+
+
+
+::: challenge
+
+# Plot line width by attribute
+
+In the example above, we set the line widths to be 1, 0.75, 0.5, and 0.25. In our case line thicknesses are consistent with the hierarchy of the selected road types, but in some cases we might want to show a different hierarchy.
+
+Let’s create another plot where we show the different line types with the following thicknesses:
+
+- motorways linewidth = 0.25
+- primary linewidth = 0.75
+- secondary linewidth = 0.5
+- cycleway linewidth = 1
+
+::: solution
+
+
+```r
+levels(factor(lines_Delft_selection$highway))
+```
+
+```{.output}
+[1] "motorway" "primary" "secondary" "cycleway"
+```
+
+
+```r
+line_width <- c(0.25, 0.75, 0.5, 1)
+```
+
+
+```r
+ggplot(data = lines_Delft_selection) +
+ geom_sf(aes(linewidth = highway)) +
+ scale_linewidth_manual(values = line_width) +
+ labs(title = "Mobility network of Delft",
+ subtitle = "Roads & Cycleways - Line width varies") +
+ coord_sf(datum = st_crs(28992))
+```
+
+ +
+:::
+
+:::
+
+
+## Add plot legend
+
+Let’s add a legend to our plot. We will use the `road_colors` object that we created above to color the legend. We can customize the appearance of our legend by manually setting different parameters.
+
+
+```r
+ggplot(data = lines_Delft_selection) +
+ geom_sf(aes(color = highway), linewidth = 1.5) +
+ scale_color_manual(values = road_colors) +
+ labs(color = 'Road Type') +
+ labs(title = "Mobility network of Delft",
+ subtitle = "Roads & Cycleways - Default Legend") +
+ coord_sf(datum = st_crs(28992))
+```
+
+
+
+:::
+
+:::
+
+
+## Add plot legend
+
+Let’s add a legend to our plot. We will use the `road_colors` object that we created above to color the legend. We can customize the appearance of our legend by manually setting different parameters.
+
+
+```r
+ggplot(data = lines_Delft_selection) +
+ geom_sf(aes(color = highway), linewidth = 1.5) +
+ scale_color_manual(values = road_colors) +
+ labs(color = 'Road Type') +
+ labs(title = "Mobility network of Delft",
+ subtitle = "Roads & Cycleways - Default Legend") +
+ coord_sf(datum = st_crs(28992))
+```
+
+ +
+Mobility network in Delft using thicker lines than the previous example.
+ +
+Map of the mobility network in Delft with large-font and border around the legend.
+ +
+Map of the mobility network in Delft using a different color palette.
+ +
+:::
+
+:::
+
+::: challenge
+
+# Plot polygon by attribute
+
+
+Create a map of the municipal boundaries in the Netherlands using the data located in your data folder: `nl-gemeenten.shp`. Apply a line color to each state using its region value. Add a legend.
+
+::: solution
+
+
+```r
+municipal_boundaries_NL <- st_read(here("data", "nl-gemeenten.shp"))
+```
+
+```{.output}
+Reading layer `nl-gemeenten' from data source
+ `/home/runner/work/r-geospatial-urban/r-geospatial-urban/site/built/data/nl-gemeenten.shp'
+ using driver `ESRI Shapefile'
+Simple feature collection with 344 features and 6 fields
+Geometry type: MULTIPOLYGON
+Dimension: XY
+Bounding box: xmin: 10425.16 ymin: 306846.2 xmax: 278026.1 ymax: 621876.3
+Projected CRS: Amersfoort / RD New
+```
+
+
+```r
+str(municipal_boundaries_NL)
+```
+
+```{.output}
+Classes 'sf' and 'data.frame': 344 obs. of 7 variables:
+ $ identifica: chr "GM0014" "GM0034" "GM0037" "GM0047" ...
+ $ naam : chr "Groningen" "Almere" "Stadskanaal" "Veendam" ...
+ $ code : chr "0014" "0034" "0037" "0047" ...
+ $ ligtInProv: chr "20" "24" "20" "20" ...
+ $ ligtInPr_1: chr "Groningen" "Flevoland" "Groningen" "Groningen" ...
+ $ fuuid : chr "gemeentegebied.ee21436e-5a2d-4a8f-b2bf-113bddd028fc" "gemeentegebied.6e4378d7-0905-4dff-b351-57c1940c9c90" "gemeentegebied.515fbfe4-614e-463d-8b8c-91d35ca93b3b" "gemeentegebied.a3e71341-218c-44bf-ba12-01e2251ea2f6" ...
+ $ geometry :sfc_MULTIPOLYGON of length 344; first list element: List of 1
+ ..$ :List of 1
+ .. ..$ : num [1:4749, 1:2] 238742 238741 238740 238738 238735 ...
+ ..- attr(*, "class")= chr [1:3] "XY" "MULTIPOLYGON" "sfg"
+ - attr(*, "sf_column")= chr "geometry"
+ - attr(*, "agr")= Factor w/ 3 levels "constant","aggregate",..: NA NA NA NA NA NA
+ ..- attr(*, "names")= chr [1:6] "identifica" "naam" "code" "ligtInProv" ...
+```
+
+```r
+levels(factor(municipal_boundaries_NL$ligtInPr_1))
+```
+
+```{.output}
+ [1] "Drenthe" "Flevoland" "Fryslân" "Gelderland"
+ [5] "Groningen" "Limburg" "Noord-Brabant" "Noord-Holland"
+ [9] "Overijssel" "Utrecht" "Zeeland" "Zuid-Holland"
+```
+
+
+```r
+ggplot(data = municipal_boundaries_NL) +
+ geom_sf(aes(color = ligtInPr_1), linewidth = 1) +
+ labs(title = "Contiguous NL Municipal Boundaries") +
+ coord_sf(datum = st_crs(28992))
+```
+
+
+
+:::
+
+:::
+
+::: challenge
+
+# Plot polygon by attribute
+
+
+Create a map of the municipal boundaries in the Netherlands using the data located in your data folder: `nl-gemeenten.shp`. Apply a line color to each state using its region value. Add a legend.
+
+::: solution
+
+
+```r
+municipal_boundaries_NL <- st_read(here("data", "nl-gemeenten.shp"))
+```
+
+```{.output}
+Reading layer `nl-gemeenten' from data source
+ `/home/runner/work/r-geospatial-urban/r-geospatial-urban/site/built/data/nl-gemeenten.shp'
+ using driver `ESRI Shapefile'
+Simple feature collection with 344 features and 6 fields
+Geometry type: MULTIPOLYGON
+Dimension: XY
+Bounding box: xmin: 10425.16 ymin: 306846.2 xmax: 278026.1 ymax: 621876.3
+Projected CRS: Amersfoort / RD New
+```
+
+
+```r
+str(municipal_boundaries_NL)
+```
+
+```{.output}
+Classes 'sf' and 'data.frame': 344 obs. of 7 variables:
+ $ identifica: chr "GM0014" "GM0034" "GM0037" "GM0047" ...
+ $ naam : chr "Groningen" "Almere" "Stadskanaal" "Veendam" ...
+ $ code : chr "0014" "0034" "0037" "0047" ...
+ $ ligtInProv: chr "20" "24" "20" "20" ...
+ $ ligtInPr_1: chr "Groningen" "Flevoland" "Groningen" "Groningen" ...
+ $ fuuid : chr "gemeentegebied.ee21436e-5a2d-4a8f-b2bf-113bddd028fc" "gemeentegebied.6e4378d7-0905-4dff-b351-57c1940c9c90" "gemeentegebied.515fbfe4-614e-463d-8b8c-91d35ca93b3b" "gemeentegebied.a3e71341-218c-44bf-ba12-01e2251ea2f6" ...
+ $ geometry :sfc_MULTIPOLYGON of length 344; first list element: List of 1
+ ..$ :List of 1
+ .. ..$ : num [1:4749, 1:2] 238742 238741 238740 238738 238735 ...
+ ..- attr(*, "class")= chr [1:3] "XY" "MULTIPOLYGON" "sfg"
+ - attr(*, "sf_column")= chr "geometry"
+ - attr(*, "agr")= Factor w/ 3 levels "constant","aggregate",..: NA NA NA NA NA NA
+ ..- attr(*, "names")= chr [1:6] "identifica" "naam" "code" "ligtInProv" ...
+```
+
+```r
+levels(factor(municipal_boundaries_NL$ligtInPr_1))
+```
+
+```{.output}
+ [1] "Drenthe" "Flevoland" "Fryslân" "Gelderland"
+ [5] "Groningen" "Limburg" "Noord-Brabant" "Noord-Holland"
+ [9] "Overijssel" "Utrecht" "Zeeland" "Zuid-Holland"
+```
+
+
+```r
+ggplot(data = municipal_boundaries_NL) +
+ geom_sf(aes(color = ligtInPr_1), linewidth = 1) +
+ labs(title = "Contiguous NL Municipal Boundaries") +
+ coord_sf(datum = st_crs(28992))
+```
+
+ +
+:::
+
+:::
+
+
+::::::::::::::::::::::::::::::::::::: keypoints
+
+- Spatial objects in `sf` are similar to standard data frames and can be manipulated using the same functions.
+
+- Almost any feature of a plot can be customized using the various functions and options in the `ggplot2` package.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
diff --git a/11-plot-multiple-shape-files.md b/11-plot-multiple-shape-files.md
new file mode 100644
index 00000000..826bc823
--- /dev/null
+++ b/11-plot-multiple-shape-files.md
@@ -0,0 +1,172 @@
+---
+title: 'Plot multiple shapefiles'
+teaching: 40
+exercises: 20
+---
+
+
+
+:::::::::::::::::::::::::::::::::::::: questions
+
+- How can I create map compositions with custom legends using ggplot?
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::::::::::::::::::::::::::::::::::::: objectives
+
+- Plot multiple vector layers in the same plot.
+- Apply custom symbols to spatial objects in a plot.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+This episode builds upon the [previous episode]() to work with vector layers in R and explore how to plot multiple vector layers.
+
+
+
+## Load the data
+
+To work with vector data in R, we can use the `sf` library. The `terra` package also allows us to explore metadata using similar commands for both raster and vector files. Make sure that you have these packages loaded.
+
+We will continue to work with the three ESRI shapefiles that we loaded in the [Open and Plot Vector Layers]() episode.
+
+
+
+## Plotting Multiple Vector Layers
+
+So far we learned how to plot information from a single shapefile and do some plot customization. What if we want to create a more complex plot with many shapefiles and unique symbols that need to be represented clearly in a legend?
+
+We will create a plot that combines our leisure locations (`point_Delft`), municipal boundary (`boundary_Delft`) and streets (`lines_Delft`) spatial objects. We will need to build a custom legend as well.
+
+To begin, we will create a plot with the site boundary as the first layer. Then layer the leisure locations and street data on top using `+`.
+
+
+```r
+ggplot() +
+ geom_sf(data = boundary_Delft, fill = "lightgrey", color = "lightgrey") +
+ geom_sf(data = lines_Delft_selection, aes(color = highway), size = 1) +
+ geom_sf(data = point_Delft) +
+ labs(title = "Mobility network of Delft") +
+ coord_sf(datum = st_crs(28992))
+```
+
+
+
+:::
+
+:::
+
+
+::::::::::::::::::::::::::::::::::::: keypoints
+
+- Spatial objects in `sf` are similar to standard data frames and can be manipulated using the same functions.
+
+- Almost any feature of a plot can be customized using the various functions and options in the `ggplot2` package.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
diff --git a/11-plot-multiple-shape-files.md b/11-plot-multiple-shape-files.md
new file mode 100644
index 00000000..826bc823
--- /dev/null
+++ b/11-plot-multiple-shape-files.md
@@ -0,0 +1,172 @@
+---
+title: 'Plot multiple shapefiles'
+teaching: 40
+exercises: 20
+---
+
+
+
+:::::::::::::::::::::::::::::::::::::: questions
+
+- How can I create map compositions with custom legends using ggplot?
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::::::::::::::::::::::::::::::::::::: objectives
+
+- Plot multiple vector layers in the same plot.
+- Apply custom symbols to spatial objects in a plot.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+This episode builds upon the [previous episode]() to work with vector layers in R and explore how to plot multiple vector layers.
+
+
+
+## Load the data
+
+To work with vector data in R, we can use the `sf` library. The `terra` package also allows us to explore metadata using similar commands for both raster and vector files. Make sure that you have these packages loaded.
+
+We will continue to work with the three ESRI shapefiles that we loaded in the [Open and Plot Vector Layers]() episode.
+
+
+
+## Plotting Multiple Vector Layers
+
+So far we learned how to plot information from a single shapefile and do some plot customization. What if we want to create a more complex plot with many shapefiles and unique symbols that need to be represented clearly in a legend?
+
+We will create a plot that combines our leisure locations (`point_Delft`), municipal boundary (`boundary_Delft`) and streets (`lines_Delft`) spatial objects. We will need to build a custom legend as well.
+
+To begin, we will create a plot with the site boundary as the first layer. Then layer the leisure locations and street data on top using `+`.
+
+
+```r
+ggplot() +
+ geom_sf(data = boundary_Delft, fill = "lightgrey", color = "lightgrey") +
+ geom_sf(data = lines_Delft_selection, aes(color = highway), size = 1) +
+ geom_sf(data = point_Delft) +
+ labs(title = "Mobility network of Delft") +
+ coord_sf(datum = st_crs(28992))
+```
+
+ +
+Next, let’s build a custom legend using the symbology (the colors and symbols) that we used to create the plot above. For example, it might be good if the lines were symbolized as lines. In the previous episode, you may have noticed that the default legend behavior for `geom_sf` is to draw a ‘patch’ for each legend entry. If you want the legend to draw lines or points, you need to add an instruction to the `geom_sf` call - in this case, `show.legend = 'line'`.
+
+
+```r
+leisure_colors <- rainbow(15)
+point_Delft$leisure <- factor(point_Delft$leisure)
+ggplot() +
+ geom_sf(data = boundary_Delft, fill = "lightgrey", color = "lightgrey") +
+ geom_sf(data = lines_Delft_selection, aes(color = highway), size = 1) +
+ geom_sf(data = point_Delft, aes(fill = leisure), shape = 21) +
+ scale_color_manual(values = road_colors, name = "Road Type") +
+ scale_fill_manual(values = leisure_colors, name = "Lesiure Location") +
+ labs(title = "Mobility network and leisure in Delft") +
+ coord_sf(datum = st_crs(28992))
+```
+
+
+
+Next, let’s build a custom legend using the symbology (the colors and symbols) that we used to create the plot above. For example, it might be good if the lines were symbolized as lines. In the previous episode, you may have noticed that the default legend behavior for `geom_sf` is to draw a ‘patch’ for each legend entry. If you want the legend to draw lines or points, you need to add an instruction to the `geom_sf` call - in this case, `show.legend = 'line'`.
+
+
+```r
+leisure_colors <- rainbow(15)
+point_Delft$leisure <- factor(point_Delft$leisure)
+ggplot() +
+ geom_sf(data = boundary_Delft, fill = "lightgrey", color = "lightgrey") +
+ geom_sf(data = lines_Delft_selection, aes(color = highway), size = 1) +
+ geom_sf(data = point_Delft, aes(fill = leisure), shape = 21) +
+ scale_color_manual(values = road_colors, name = "Road Type") +
+ scale_fill_manual(values = leisure_colors, name = "Lesiure Location") +
+ labs(title = "Mobility network and leisure in Delft") +
+ coord_sf(datum = st_crs(28992))
+```
+
+ +
+
+```r
+ggplot() +
+ geom_sf(data = boundary_Delft, fill = "lightgrey", color = "lightgrey") +
+ geom_sf(data = lines_Delft_selection, aes(color = highway), size = 1) +
+ geom_sf(data = point_Delft, aes(fill = leisure), shape = 22) +
+ scale_color_manual(values = road_colors, name = "Line Type") +
+ scale_fill_manual(values = leisure_colors, name = "Leisure Location") +
+ labs(title = "Mobility network and leisure in Delft") +
+ coord_sf(datum = st_crs(28992))
+```
+
+
+
+
+```r
+ggplot() +
+ geom_sf(data = boundary_Delft, fill = "lightgrey", color = "lightgrey") +
+ geom_sf(data = lines_Delft_selection, aes(color = highway), size = 1) +
+ geom_sf(data = point_Delft, aes(fill = leisure), shape = 22) +
+ scale_color_manual(values = road_colors, name = "Line Type") +
+ scale_fill_manual(values = leisure_colors, name = "Leisure Location") +
+ labs(title = "Mobility network and leisure in Delft") +
+ coord_sf(datum = st_crs(28992))
+```
+
+ +
+We notice that there are quite some playgrounds in the residential parts of Delft, whereas on campus there is a concentration of picnic tables. So that is what our next challenge is about.
+
+### Challenge 7 (5 minutes)
+
+Create a map of leisure locations only including `playground` and `picnic_table`, with each point colored by the leisure type. Overlay this layer on top of the `lines_Delft` layer (the streets). Create a custom legend that applies line symbols to lines and point symbols to the points.
+
+Modify the plot above. Tell R to plot each point, using a different symbol of shape value.
+
+
+```r
+leisure_locations_selection <- st_read("data/delft-leisure.shp") %>%
+ filter(leisure %in% c("playground", "picnic_table"))
+```
+
+```{.output}
+Reading layer `delft-leisure' from data source
+ `/home/runner/work/r-geospatial-urban/r-geospatial-urban/site/built/data/delft-leisure.shp'
+ using driver `ESRI Shapefile'
+Simple feature collection with 298 features and 2 fields
+Geometry type: POINT
+Dimension: XY
+Bounding box: xmin: 81863.21 ymin: 442621.1 xmax: 87370.15 ymax: 449345.1
+Projected CRS: Amersfoort / RD New
+```
+
+
+```r
+levels(factor(leisure_locations_selection$leisure))
+```
+
+```{.output}
+[1] "picnic_table" "playground"
+```
+
+
+```r
+blue_orange <- c("cornflowerblue", "darkorange")
+```
+
+
+```r
+ggplot() +
+ geom_sf(data = lines_Delft_selection, aes(color = highway)) +
+ geom_sf(data = leisure_locations_selection, aes(fill = leisure),
+ shape = 21, show.legend = 'point') +
+ scale_color_manual(name = "Line Type", values = road_colors,
+ guide = guide_legend(override.aes = list(linetype = "solid", shape = NA))) +
+ scale_fill_manual(name = "Soil Type", values = blue_orange,
+ guide = guide_legend(override.aes = list(linetype = "blank", shape = 21, colour = NA))) +
+ labs(title = "Traffic and leisure") +
+ coord_sf(datum = st_crs(28992))
+```
+
+
+
+We notice that there are quite some playgrounds in the residential parts of Delft, whereas on campus there is a concentration of picnic tables. So that is what our next challenge is about.
+
+### Challenge 7 (5 minutes)
+
+Create a map of leisure locations only including `playground` and `picnic_table`, with each point colored by the leisure type. Overlay this layer on top of the `lines_Delft` layer (the streets). Create a custom legend that applies line symbols to lines and point symbols to the points.
+
+Modify the plot above. Tell R to plot each point, using a different symbol of shape value.
+
+
+```r
+leisure_locations_selection <- st_read("data/delft-leisure.shp") %>%
+ filter(leisure %in% c("playground", "picnic_table"))
+```
+
+```{.output}
+Reading layer `delft-leisure' from data source
+ `/home/runner/work/r-geospatial-urban/r-geospatial-urban/site/built/data/delft-leisure.shp'
+ using driver `ESRI Shapefile'
+Simple feature collection with 298 features and 2 fields
+Geometry type: POINT
+Dimension: XY
+Bounding box: xmin: 81863.21 ymin: 442621.1 xmax: 87370.15 ymax: 449345.1
+Projected CRS: Amersfoort / RD New
+```
+
+
+```r
+levels(factor(leisure_locations_selection$leisure))
+```
+
+```{.output}
+[1] "picnic_table" "playground"
+```
+
+
+```r
+blue_orange <- c("cornflowerblue", "darkorange")
+```
+
+
+```r
+ggplot() +
+ geom_sf(data = lines_Delft_selection, aes(color = highway)) +
+ geom_sf(data = leisure_locations_selection, aes(fill = leisure),
+ shape = 21, show.legend = 'point') +
+ scale_color_manual(name = "Line Type", values = road_colors,
+ guide = guide_legend(override.aes = list(linetype = "solid", shape = NA))) +
+ scale_fill_manual(name = "Soil Type", values = blue_orange,
+ guide = guide_legend(override.aes = list(linetype = "blank", shape = 21, colour = NA))) +
+ labs(title = "Traffic and leisure") +
+ coord_sf(datum = st_crs(28992))
+```
+
+ +
+
+```r
+ggplot() +
+ geom_sf(data = lines_Delft_selection, aes(color = highway), size = 1) +
+ geom_sf(data = leisure_locations_selection, aes(fill = leisure, shape = leisure), size = 2) +
+ scale_shape_manual(name = "Leisure Type", values = c(21, 22)) +
+ scale_color_manual(name = "Line Type", values = road_colors) +
+ scale_fill_manual(name = "Leisure Type", values = rainbow(15),
+ guide = guide_legend(override.aes = list(linetype = "blank", shape = c(21, 22),
+ color = "black"))) +
+ labs(title = "Road network and leisure") +
+ coord_sf(datum = st_crs(28992))
+```
+
+
+
+
+```r
+ggplot() +
+ geom_sf(data = lines_Delft_selection, aes(color = highway), size = 1) +
+ geom_sf(data = leisure_locations_selection, aes(fill = leisure, shape = leisure), size = 2) +
+ scale_shape_manual(name = "Leisure Type", values = c(21, 22)) +
+ scale_color_manual(name = "Line Type", values = road_colors) +
+ scale_fill_manual(name = "Leisure Type", values = rainbow(15),
+ guide = guide_legend(override.aes = list(linetype = "blank", shape = c(21, 22),
+ color = "black"))) +
+ labs(title = "Road network and leisure") +
+ coord_sf(datum = st_crs(28992))
+```
+
+ +
+
+
+
+
+
+
+
+::::::::::::::::::::::::::::::::::::: keypoints
+
+- Use the `+` operator to add multiple layers to a ggplot.
+- A plot can be a combination of multiple vector layers.
+- Use the `show.legend` argument to set legend symbol types.
+- Use the `scale_fill_manual()` function to set legend colors.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
diff --git a/12-handling-spatial-projection-and-crs.md b/12-handling-spatial-projection-and-crs.md
new file mode 100644
index 00000000..8dce6344
--- /dev/null
+++ b/12-handling-spatial-projection-and-crs.md
@@ -0,0 +1,200 @@
+---
+title: 'Handling Spatial Projections & CRS'
+teaching: 40
+exercises: 20
+---
+
+
+
+:::::::::::::::::::::::::::::::::::::: questions
+
+- What do I do when vector data don’t line up?
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::::::::::::::::::::::::::::::::::::: objectives
+
+- Plot vector objects with different CRSs in the same plot.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+## Working with spatial data from different sources
+
+
+```r
+municipal_boundary_NL <- st_read("data/nl-gemeenten.shp")
+```
+
+```{.output}
+Reading layer `nl-gemeenten' from data source
+ `/home/runner/work/r-geospatial-urban/r-geospatial-urban/site/built/data/nl-gemeenten.shp'
+ using driver `ESRI Shapefile'
+Simple feature collection with 344 features and 6 fields
+Geometry type: MULTIPOLYGON
+Dimension: XY
+Bounding box: xmin: 10425.16 ymin: 306846.2 xmax: 278026.1 ymax: 621876.3
+Projected CRS: Amersfoort / RD New
+```
+
+
+```r
+ggplot() +
+ geom_sf(data = municipal_boundary_NL) +
+ labs(title = "Map of Contiguous NL Municipal Boundaries") +
+ coord_sf(datum = st_crs(28992))
+```
+
+
+
+
+
+
+
+
+
+
+::::::::::::::::::::::::::::::::::::: keypoints
+
+- Use the `+` operator to add multiple layers to a ggplot.
+- A plot can be a combination of multiple vector layers.
+- Use the `show.legend` argument to set legend symbol types.
+- Use the `scale_fill_manual()` function to set legend colors.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
diff --git a/12-handling-spatial-projection-and-crs.md b/12-handling-spatial-projection-and-crs.md
new file mode 100644
index 00000000..8dce6344
--- /dev/null
+++ b/12-handling-spatial-projection-and-crs.md
@@ -0,0 +1,200 @@
+---
+title: 'Handling Spatial Projections & CRS'
+teaching: 40
+exercises: 20
+---
+
+
+
+:::::::::::::::::::::::::::::::::::::: questions
+
+- What do I do when vector data don’t line up?
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::::::::::::::::::::::::::::::::::::: objectives
+
+- Plot vector objects with different CRSs in the same plot.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+## Working with spatial data from different sources
+
+
+```r
+municipal_boundary_NL <- st_read("data/nl-gemeenten.shp")
+```
+
+```{.output}
+Reading layer `nl-gemeenten' from data source
+ `/home/runner/work/r-geospatial-urban/r-geospatial-urban/site/built/data/nl-gemeenten.shp'
+ using driver `ESRI Shapefile'
+Simple feature collection with 344 features and 6 fields
+Geometry type: MULTIPOLYGON
+Dimension: XY
+Bounding box: xmin: 10425.16 ymin: 306846.2 xmax: 278026.1 ymax: 621876.3
+Projected CRS: Amersfoort / RD New
+```
+
+
+```r
+ggplot() +
+ geom_sf(data = municipal_boundary_NL) +
+ labs(title = "Map of Contiguous NL Municipal Boundaries") +
+ coord_sf(datum = st_crs(28992))
+```
+
+ +
+We can add a country boundary layer to make it look nicer. If we specify a thicker line width using size = 2 for the country boundary layer, it will make our map pop!
+
+
+```r
+country_boundary_NL <- st_read("data/nl-boundary.shp")
+```
+
+```{.output}
+Reading layer `nl-boundary' from data source
+ `/home/runner/work/r-geospatial-urban/r-geospatial-urban/site/built/data/nl-boundary.shp'
+ using driver `ESRI Shapefile'
+Simple feature collection with 1 feature and 1 field
+Geometry type: MULTIPOLYGON
+Dimension: XY
+Bounding box: xmin: 10425.16 ymin: 306846.2 xmax: 278026.1 ymax: 621876.3
+Projected CRS: Amersfoort / RD New
+```
+
+
+```r
+ggplot() +
+ geom_sf(data = country_boundary_NL, color = "gray18", linewidth = 2) +
+ geom_sf(data = municipal_boundary_NL, color = "gray40") +
+ labs(title = "Map of Contiguous NL Municipal Boundaries") +
+ coord_sf(datum = st_crs(28992))
+```
+
+
+
+We can add a country boundary layer to make it look nicer. If we specify a thicker line width using size = 2 for the country boundary layer, it will make our map pop!
+
+
+```r
+country_boundary_NL <- st_read("data/nl-boundary.shp")
+```
+
+```{.output}
+Reading layer `nl-boundary' from data source
+ `/home/runner/work/r-geospatial-urban/r-geospatial-urban/site/built/data/nl-boundary.shp'
+ using driver `ESRI Shapefile'
+Simple feature collection with 1 feature and 1 field
+Geometry type: MULTIPOLYGON
+Dimension: XY
+Bounding box: xmin: 10425.16 ymin: 306846.2 xmax: 278026.1 ymax: 621876.3
+Projected CRS: Amersfoort / RD New
+```
+
+
+```r
+ggplot() +
+ geom_sf(data = country_boundary_NL, color = "gray18", linewidth = 2) +
+ geom_sf(data = municipal_boundary_NL, color = "gray40") +
+ labs(title = "Map of Contiguous NL Municipal Boundaries") +
+ coord_sf(datum = st_crs(28992))
+```
+
+ +
+
+```r
+# st_crs(point_Delft)
+```
+
+
+```r
+st_crs(municipal_boundary_NL)$epsg
+```
+
+```{.output}
+[1] 28992
+```
+
+
+```r
+st_crs(country_boundary_NL)$epsg
+```
+
+```{.output}
+[1] 28992
+```
+
+
+```r
+boundary_Delft <- st_read("data/delft-boundary.shp")
+```
+
+```{.output}
+Reading layer `delft-boundary' from data source
+ `/home/runner/work/r-geospatial-urban/r-geospatial-urban/site/built/data/delft-boundary.shp'
+ using driver `ESRI Shapefile'
+Simple feature collection with 1 feature and 1 field
+Geometry type: POLYGON
+Dimension: XY
+Bounding box: xmin: 4.320218 ymin: 51.96632 xmax: 4.407911 ymax: 52.0326
+Geodetic CRS: WGS 84
+```
+
+```r
+st_crs(boundary_Delft)$epsg
+```
+
+```{.output}
+[1] 4326
+```
+
+```r
+boundary_Delft <- st_transform(boundary_Delft, 28992)
+```
+
+
+```r
+ggplot() +
+ geom_sf(data = country_boundary_NL, linewidth = 2, color = "gray18") +
+ geom_sf(data = municipal_boundary_NL, color = "gray40") +
+ geom_sf(data = boundary_Delft, color = "purple", fill = "purple") +
+ labs(title = "Map of Contiguous NL Municipal Boundaries") +
+ coord_sf(datum = st_crs(28992))
+```
+
+
+
+
+```r
+# st_crs(point_Delft)
+```
+
+
+```r
+st_crs(municipal_boundary_NL)$epsg
+```
+
+```{.output}
+[1] 28992
+```
+
+
+```r
+st_crs(country_boundary_NL)$epsg
+```
+
+```{.output}
+[1] 28992
+```
+
+
+```r
+boundary_Delft <- st_read("data/delft-boundary.shp")
+```
+
+```{.output}
+Reading layer `delft-boundary' from data source
+ `/home/runner/work/r-geospatial-urban/r-geospatial-urban/site/built/data/delft-boundary.shp'
+ using driver `ESRI Shapefile'
+Simple feature collection with 1 feature and 1 field
+Geometry type: POLYGON
+Dimension: XY
+Bounding box: xmin: 4.320218 ymin: 51.96632 xmax: 4.407911 ymax: 52.0326
+Geodetic CRS: WGS 84
+```
+
+```r
+st_crs(boundary_Delft)$epsg
+```
+
+```{.output}
+[1] 4326
+```
+
+```r
+boundary_Delft <- st_transform(boundary_Delft, 28992)
+```
+
+
+```r
+ggplot() +
+ geom_sf(data = country_boundary_NL, linewidth = 2, color = "gray18") +
+ geom_sf(data = municipal_boundary_NL, color = "gray40") +
+ geom_sf(data = boundary_Delft, color = "purple", fill = "purple") +
+ labs(title = "Map of Contiguous NL Municipal Boundaries") +
+ coord_sf(datum = st_crs(28992))
+```
+
+ +
+::: challenge
+
+# Plot multiple layers of spatial data
+
+
+Create a map of South Holland as follows:
+
+1. Import and plot `nl-gemeenten.shp`. Adjust line width as necessary.
+2. Layer the boundary of Delft onto the plot.
+3. Add a title.
+4. Add a legend that shows both the province boundaries (as a line) and the boundary of Delft (as a filled polygon).
+
+::: solution
+
+
+```r
+boundary_ZH <- municipal_boundary_NL %>%
+ filter(ligtInPr_1 == "Zuid-Holland")
+```
+
+
+```r
+ggplot() +
+ geom_sf(data = boundary_ZH, aes(color ="color"), show.legend = "line") +
+ scale_color_manual(name = "", labels = "Municipal Boundaries in South Holland", values = c("color" = "gray18")) +
+ geom_sf(data = boundary_Delft, aes(shape = "shape"), color = "purple", fill = "purple") +
+ scale_shape_manual(name = "", labels = "Municipality of Delft", values = c("shape" = 19)) +
+ labs(title = "Delft location") +
+ theme(legend.background = element_rect(color = NA)) +
+ coord_sf(datum = st_crs(28992))
+```
+
+
+
+::: challenge
+
+# Plot multiple layers of spatial data
+
+
+Create a map of South Holland as follows:
+
+1. Import and plot `nl-gemeenten.shp`. Adjust line width as necessary.
+2. Layer the boundary of Delft onto the plot.
+3. Add a title.
+4. Add a legend that shows both the province boundaries (as a line) and the boundary of Delft (as a filled polygon).
+
+::: solution
+
+
+```r
+boundary_ZH <- municipal_boundary_NL %>%
+ filter(ligtInPr_1 == "Zuid-Holland")
+```
+
+
+```r
+ggplot() +
+ geom_sf(data = boundary_ZH, aes(color ="color"), show.legend = "line") +
+ scale_color_manual(name = "", labels = "Municipal Boundaries in South Holland", values = c("color" = "gray18")) +
+ geom_sf(data = boundary_Delft, aes(shape = "shape"), color = "purple", fill = "purple") +
+ scale_shape_manual(name = "", labels = "Municipality of Delft", values = c("shape" = 19)) +
+ labs(title = "Delft location") +
+ theme(legend.background = element_rect(color = NA)) +
+ coord_sf(datum = st_crs(28992))
+```
+
+ +
+:::
+
+:::
+
+
+
+## Export a shapefile
+
+To save a file, use the `st_write()` function from the `sf` package.
+
+
+```r
+st_write(leisure_locations_selection,
+ "data/leisure_locations_selection.shp"), driver = "ESRI Shapefile")
+```
+
+
+
+::::::::::::::::::::::::::::::::::::: keypoints
+
+- `ggplot2` automatically converts all objects in a plot to the same CRS.
+- Still be aware of the CRS and extent for each object.
+- You can export an `sf` object to a shapefile with `st_write()`.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
diff --git a/13-intro-to-raster-data.md b/13-intro-to-raster-data.md
new file mode 100644
index 00000000..c0ce105c
--- /dev/null
+++ b/13-intro-to-raster-data.md
@@ -0,0 +1,478 @@
+---
+title: 'Intro to Raster Data'
+teaching: 30
+exercises: 2
+---
+
+
+
+::: questions
+- What is a raster dataset?
+- How do I work with and plot raster data in R?
+:::
+
+::: objectives
+
+After completing this episode, participants should be able to…
+
+- Explore raster attributes and metadata using R.
+- Import rasters into R using the `terra` package.
+- Plot a raster file in R using the `ggplot2` package.
+- Describe the difference between single- and multi-band rasters.
+
+:::
+
+::: prereq
+
+# Things you'll need to complete this episode
+
+See the [setup instructions](../learners/setup.md) for detailed information about the software, data, and other prerequisites you will need to work through the examples in this episode.
+
+
+
+:::
+
+In this lesson, we will work with raster data. We will start with an introduction of the fundamental principles and metadata needed to work with raster data in R. We will discuss some of the core metadata elements needed to understand raster data in R, including CRS and resolution.
+
+We continue to work with the `tidyverse` package and we will use the `terra` package to work with raster data. Make sure that you have those packages loaded.
+
+
+```r
+library(tidyverse)
+library(terra)
+```
+
+::: callout
+
+# The data used in this lesson
+
+In this and lesson, we will use:
+
+- data extracted from the [AHN digital elevation dataset of the Netherlands](https://www.ahn.nl/) for the TU Delft campus area; and
+- high-resolution RGB aerial photos of the TU Delft library obtained from [Beeldmateriaal Nederland](https://www.beeldmateriaal.nl/download-luchtfotos).
+
+:::
+
+## View Raster File Attributes
+
+We will be working with a series of GeoTIFF files in this lesson. The GeoTIFF format contains a set of embedded tags with metadata about the raster data. We can use the function `describe()` from the `terra` package to get information about our raster data before we read that data into R. It is recommended to do this before importing your data. We first examine the file `tud-dsm-5m.tif`.
+
+
+```r
+describe("data/tud-dsm-5m.tif")
+```
+
+```{.output}
+ [1] "Driver: GTiff/GeoTIFF"
+ [2] "Files: data/tud-dsm-5m.tif"
+ [3] "Size is 722, 386"
+ [4] "Coordinate System is:"
+ [5] "PROJCRS[\"Amersfoort / RD New\","
+ [6] " BASEGEOGCRS[\"Amersfoort\","
+ [7] " DATUM[\"Amersfoort\","
+ [8] " ELLIPSOID[\"Bessel 1841\",6377397.155,299.1528128,"
+ [9] " LENGTHUNIT[\"metre\",1]]],"
+[10] " PRIMEM[\"Greenwich\",0,"
+[11] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+[12] " ID[\"EPSG\",4289]],"
+[13] " CONVERSION[\"RD New\","
+[14] " METHOD[\"Oblique Stereographic\","
+[15] " ID[\"EPSG\",9809]],"
+[16] " PARAMETER[\"Latitude of natural origin\",52.1561605555556,"
+[17] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[18] " ID[\"EPSG\",8801]],"
+[19] " PARAMETER[\"Longitude of natural origin\",5.38763888888889,"
+[20] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[21] " ID[\"EPSG\",8802]],"
+[22] " PARAMETER[\"Scale factor at natural origin\",0.9999079,"
+[23] " SCALEUNIT[\"unity\",1],"
+[24] " ID[\"EPSG\",8805]],"
+[25] " PARAMETER[\"False easting\",155000,"
+[26] " LENGTHUNIT[\"metre\",1],"
+[27] " ID[\"EPSG\",8806]],"
+[28] " PARAMETER[\"False northing\",463000,"
+[29] " LENGTHUNIT[\"metre\",1],"
+[30] " ID[\"EPSG\",8807]]],"
+[31] " CS[Cartesian,2],"
+[32] " AXIS[\"easting (X)\",east,"
+[33] " ORDER[1],"
+[34] " LENGTHUNIT[\"metre\",1]],"
+[35] " AXIS[\"northing (Y)\",north,"
+[36] " ORDER[2],"
+[37] " LENGTHUNIT[\"metre\",1]],"
+[38] " USAGE["
+[39] " SCOPE[\"Engineering survey, topographic mapping.\"],"
+[40] " AREA[\"Netherlands - onshore, including Waddenzee, Dutch Wadden Islands and 12-mile offshore coastal zone.\"],"
+[41] " BBOX[50.75,3.2,53.7,7.22]],"
+[42] " ID[\"EPSG\",28992]]"
+[43] "Data axis to CRS axis mapping: 1,2"
+[44] "Origin = (83565.000000000000000,447180.000000000000000)"
+[45] "Pixel Size = (5.000000000000000,-5.000000000000000)"
+[46] "Metadata:"
+[47] " AREA_OR_POINT=Area"
+[48] "Image Structure Metadata:"
+[49] " INTERLEAVE=BAND"
+[50] "Corner Coordinates:"
+[51] "Upper Left ( 83565.000, 447180.000) ( 4d20'49.32\"E, 52d 0'33.67\"N)"
+[52] "Lower Left ( 83565.000, 445250.000) ( 4d20'50.77\"E, 51d59'31.22\"N)"
+[53] "Upper Right ( 87175.000, 447180.000) ( 4d23'58.60\"E, 52d 0'35.30\"N)"
+[54] "Lower Right ( 87175.000, 445250.000) ( 4d23'59.98\"E, 51d59'32.85\"N)"
+[55] "Center ( 85370.000, 446215.000) ( 4d22'24.67\"E, 52d 0' 3.27\"N)"
+[56] "Band 1 Block=722x2 Type=Float32, ColorInterp=Gray"
+```
+We will be using this information throughout this episode. By the end of the episode, you will be able to explain and understand the output above.
+
+## Open a Raster in R
+
+Now that we've previewed the metadata for our GeoTIFF, let's import this raster dataset into R and explore its metadata more closely. We can use the `rast()` function to import a raster file in R.
+
+::: callout
+# Data tip - Object names
+To improve code readability, use file and object names that make it clear what is in the file. The raster data for this episode contain the TU Delft campus and its surroundings so we’ll use a naming convention of `datatype_TUD`. The first object is a Digital Surface Model (DSM) in GeoTIFF format stored in a file `tud-dsm-5m.tif` which we will load into an object named according to our naming convention `DSM_TUD`.
+:::
+
+First we will load our raster file into R and view the data structure.
+
+
+```r
+DSM_TUD <- rast("data/tud-dsm-5m.tif")
+DSM_TUD
+```
+
+```{.output}
+class : SpatRaster
+dimensions : 386, 722, 1 (nrow, ncol, nlyr)
+resolution : 5, 5 (x, y)
+extent : 83565, 87175, 445250, 447180 (xmin, xmax, ymin, ymax)
+coord. ref. : Amersfoort / RD New (EPSG:28992)
+source : tud-dsm-5m.tif
+name : tud-dsm-5m
+```
+The information above includes a report on dimension, resolution, extent and CRS, but no information about the values. Similar to other data structures in R like vectors and data frame columns, descriptive statistics for raster data can be retrieved with the `summary()` function.
+
+
+```r
+summary(DSM_TUD)
+```
+
+```{.warning}
+Warning: [summary] used a sample
+```
+
+```{.output}
+ tud.dsm.5m
+ Min. :-5.2235
+ 1st Qu.:-0.7007
+ Median : 0.5462
+ Mean : 2.5850
+ 3rd Qu.: 4.4596
+ Max. :89.7838
+```
+
+This output gives us information about the range of values in the DSM. We can see, for instance, that the lowest elevation is `-5.2235`, the highest is `89.7838`. But note the warning. Unless you force R to calculate these statistics using every cell in the raster, it will take a random sample of 100,000 cells and calculate from them instead. To force calculation all the values, you can use the function `values`:
+
+
+```r
+summary(values(DSM_TUD))
+```
+
+```{.output}
+ tud-dsm-5m
+ Min. :-5.3907
+ 1st Qu.:-0.7008
+ Median : 0.5573
+ Mean : 2.5886
+ 3rd Qu.: 4.4648
+ Max. :92.0810
+```
+
+With a summary on all cells of the raster, the values range from a smaller minimum of `-5.3907` to a higher maximum of `92.0910`.
+
+To visualise the DSM in R using `ggplot2`, we need to convert it to a data frame. We learned about data frames in an [earlier lesson](../episodes/03-explore-data.Rmd). The `terra` package has the built-in function `as.data.frame()` for conversion to a data frame.
+
+
+```r
+DSM_TUD_df <- as.data.frame(DSM_TUD, xy = TRUE)
+```
+
+Now when we view the structure of our data, we will see a standard data frame format.
+
+
+```r
+str(DSM_TUD_df)
+```
+
+```{.output}
+'data.frame': 278692 obs. of 3 variables:
+ $ x : num 83568 83572 83578 83582 83588 ...
+ $ y : num 447178 447178 447178 447178 447178 ...
+ $ tud-dsm-5m: num 10.34 8.64 1.25 1.12 2.13 ...
+```
+
+We can use `ggplot()` to plot this data with a specific `geom_` function called `geom_raster()`. We will make the colour scale in our plot colour-blindness friendly with `scale_fill_viridis_c`, introduced in an [earlier lesson](../episodes/04-intro-to-visualisation.Rmd). We will also use the `coord_quickmap()` function to use an approximate Mercator projection for our plots. This approximation is suitable for small areas that are not too close to the poles. Other coordinate systems are available in `ggplot2` if needed, you can learn about them at their help page `?coord_map`.
+
+
+```r
+ggplot() +
+ geom_raster(data = DSM_TUD_df , aes(x = x, y = y, fill = `tud-dsm-5m`)) +
+ scale_fill_viridis_c(option = "H") + # `option = "H"` provides a contrasting colour scale
+ coord_quickmap()
+```
+
+
+
+:::
+
+:::
+
+
+
+## Export a shapefile
+
+To save a file, use the `st_write()` function from the `sf` package.
+
+
+```r
+st_write(leisure_locations_selection,
+ "data/leisure_locations_selection.shp"), driver = "ESRI Shapefile")
+```
+
+
+
+::::::::::::::::::::::::::::::::::::: keypoints
+
+- `ggplot2` automatically converts all objects in a plot to the same CRS.
+- Still be aware of the CRS and extent for each object.
+- You can export an `sf` object to a shapefile with `st_write()`.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
diff --git a/13-intro-to-raster-data.md b/13-intro-to-raster-data.md
new file mode 100644
index 00000000..c0ce105c
--- /dev/null
+++ b/13-intro-to-raster-data.md
@@ -0,0 +1,478 @@
+---
+title: 'Intro to Raster Data'
+teaching: 30
+exercises: 2
+---
+
+
+
+::: questions
+- What is a raster dataset?
+- How do I work with and plot raster data in R?
+:::
+
+::: objectives
+
+After completing this episode, participants should be able to…
+
+- Explore raster attributes and metadata using R.
+- Import rasters into R using the `terra` package.
+- Plot a raster file in R using the `ggplot2` package.
+- Describe the difference between single- and multi-band rasters.
+
+:::
+
+::: prereq
+
+# Things you'll need to complete this episode
+
+See the [setup instructions](../learners/setup.md) for detailed information about the software, data, and other prerequisites you will need to work through the examples in this episode.
+
+
+
+:::
+
+In this lesson, we will work with raster data. We will start with an introduction of the fundamental principles and metadata needed to work with raster data in R. We will discuss some of the core metadata elements needed to understand raster data in R, including CRS and resolution.
+
+We continue to work with the `tidyverse` package and we will use the `terra` package to work with raster data. Make sure that you have those packages loaded.
+
+
+```r
+library(tidyverse)
+library(terra)
+```
+
+::: callout
+
+# The data used in this lesson
+
+In this and lesson, we will use:
+
+- data extracted from the [AHN digital elevation dataset of the Netherlands](https://www.ahn.nl/) for the TU Delft campus area; and
+- high-resolution RGB aerial photos of the TU Delft library obtained from [Beeldmateriaal Nederland](https://www.beeldmateriaal.nl/download-luchtfotos).
+
+:::
+
+## View Raster File Attributes
+
+We will be working with a series of GeoTIFF files in this lesson. The GeoTIFF format contains a set of embedded tags with metadata about the raster data. We can use the function `describe()` from the `terra` package to get information about our raster data before we read that data into R. It is recommended to do this before importing your data. We first examine the file `tud-dsm-5m.tif`.
+
+
+```r
+describe("data/tud-dsm-5m.tif")
+```
+
+```{.output}
+ [1] "Driver: GTiff/GeoTIFF"
+ [2] "Files: data/tud-dsm-5m.tif"
+ [3] "Size is 722, 386"
+ [4] "Coordinate System is:"
+ [5] "PROJCRS[\"Amersfoort / RD New\","
+ [6] " BASEGEOGCRS[\"Amersfoort\","
+ [7] " DATUM[\"Amersfoort\","
+ [8] " ELLIPSOID[\"Bessel 1841\",6377397.155,299.1528128,"
+ [9] " LENGTHUNIT[\"metre\",1]]],"
+[10] " PRIMEM[\"Greenwich\",0,"
+[11] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+[12] " ID[\"EPSG\",4289]],"
+[13] " CONVERSION[\"RD New\","
+[14] " METHOD[\"Oblique Stereographic\","
+[15] " ID[\"EPSG\",9809]],"
+[16] " PARAMETER[\"Latitude of natural origin\",52.1561605555556,"
+[17] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[18] " ID[\"EPSG\",8801]],"
+[19] " PARAMETER[\"Longitude of natural origin\",5.38763888888889,"
+[20] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[21] " ID[\"EPSG\",8802]],"
+[22] " PARAMETER[\"Scale factor at natural origin\",0.9999079,"
+[23] " SCALEUNIT[\"unity\",1],"
+[24] " ID[\"EPSG\",8805]],"
+[25] " PARAMETER[\"False easting\",155000,"
+[26] " LENGTHUNIT[\"metre\",1],"
+[27] " ID[\"EPSG\",8806]],"
+[28] " PARAMETER[\"False northing\",463000,"
+[29] " LENGTHUNIT[\"metre\",1],"
+[30] " ID[\"EPSG\",8807]]],"
+[31] " CS[Cartesian,2],"
+[32] " AXIS[\"easting (X)\",east,"
+[33] " ORDER[1],"
+[34] " LENGTHUNIT[\"metre\",1]],"
+[35] " AXIS[\"northing (Y)\",north,"
+[36] " ORDER[2],"
+[37] " LENGTHUNIT[\"metre\",1]],"
+[38] " USAGE["
+[39] " SCOPE[\"Engineering survey, topographic mapping.\"],"
+[40] " AREA[\"Netherlands - onshore, including Waddenzee, Dutch Wadden Islands and 12-mile offshore coastal zone.\"],"
+[41] " BBOX[50.75,3.2,53.7,7.22]],"
+[42] " ID[\"EPSG\",28992]]"
+[43] "Data axis to CRS axis mapping: 1,2"
+[44] "Origin = (83565.000000000000000,447180.000000000000000)"
+[45] "Pixel Size = (5.000000000000000,-5.000000000000000)"
+[46] "Metadata:"
+[47] " AREA_OR_POINT=Area"
+[48] "Image Structure Metadata:"
+[49] " INTERLEAVE=BAND"
+[50] "Corner Coordinates:"
+[51] "Upper Left ( 83565.000, 447180.000) ( 4d20'49.32\"E, 52d 0'33.67\"N)"
+[52] "Lower Left ( 83565.000, 445250.000) ( 4d20'50.77\"E, 51d59'31.22\"N)"
+[53] "Upper Right ( 87175.000, 447180.000) ( 4d23'58.60\"E, 52d 0'35.30\"N)"
+[54] "Lower Right ( 87175.000, 445250.000) ( 4d23'59.98\"E, 51d59'32.85\"N)"
+[55] "Center ( 85370.000, 446215.000) ( 4d22'24.67\"E, 52d 0' 3.27\"N)"
+[56] "Band 1 Block=722x2 Type=Float32, ColorInterp=Gray"
+```
+We will be using this information throughout this episode. By the end of the episode, you will be able to explain and understand the output above.
+
+## Open a Raster in R
+
+Now that we've previewed the metadata for our GeoTIFF, let's import this raster dataset into R and explore its metadata more closely. We can use the `rast()` function to import a raster file in R.
+
+::: callout
+# Data tip - Object names
+To improve code readability, use file and object names that make it clear what is in the file. The raster data for this episode contain the TU Delft campus and its surroundings so we’ll use a naming convention of `datatype_TUD`. The first object is a Digital Surface Model (DSM) in GeoTIFF format stored in a file `tud-dsm-5m.tif` which we will load into an object named according to our naming convention `DSM_TUD`.
+:::
+
+First we will load our raster file into R and view the data structure.
+
+
+```r
+DSM_TUD <- rast("data/tud-dsm-5m.tif")
+DSM_TUD
+```
+
+```{.output}
+class : SpatRaster
+dimensions : 386, 722, 1 (nrow, ncol, nlyr)
+resolution : 5, 5 (x, y)
+extent : 83565, 87175, 445250, 447180 (xmin, xmax, ymin, ymax)
+coord. ref. : Amersfoort / RD New (EPSG:28992)
+source : tud-dsm-5m.tif
+name : tud-dsm-5m
+```
+The information above includes a report on dimension, resolution, extent and CRS, but no information about the values. Similar to other data structures in R like vectors and data frame columns, descriptive statistics for raster data can be retrieved with the `summary()` function.
+
+
+```r
+summary(DSM_TUD)
+```
+
+```{.warning}
+Warning: [summary] used a sample
+```
+
+```{.output}
+ tud.dsm.5m
+ Min. :-5.2235
+ 1st Qu.:-0.7007
+ Median : 0.5462
+ Mean : 2.5850
+ 3rd Qu.: 4.4596
+ Max. :89.7838
+```
+
+This output gives us information about the range of values in the DSM. We can see, for instance, that the lowest elevation is `-5.2235`, the highest is `89.7838`. But note the warning. Unless you force R to calculate these statistics using every cell in the raster, it will take a random sample of 100,000 cells and calculate from them instead. To force calculation all the values, you can use the function `values`:
+
+
+```r
+summary(values(DSM_TUD))
+```
+
+```{.output}
+ tud-dsm-5m
+ Min. :-5.3907
+ 1st Qu.:-0.7008
+ Median : 0.5573
+ Mean : 2.5886
+ 3rd Qu.: 4.4648
+ Max. :92.0810
+```
+
+With a summary on all cells of the raster, the values range from a smaller minimum of `-5.3907` to a higher maximum of `92.0910`.
+
+To visualise the DSM in R using `ggplot2`, we need to convert it to a data frame. We learned about data frames in an [earlier lesson](../episodes/03-explore-data.Rmd). The `terra` package has the built-in function `as.data.frame()` for conversion to a data frame.
+
+
+```r
+DSM_TUD_df <- as.data.frame(DSM_TUD, xy = TRUE)
+```
+
+Now when we view the structure of our data, we will see a standard data frame format.
+
+
+```r
+str(DSM_TUD_df)
+```
+
+```{.output}
+'data.frame': 278692 obs. of 3 variables:
+ $ x : num 83568 83572 83578 83582 83588 ...
+ $ y : num 447178 447178 447178 447178 447178 ...
+ $ tud-dsm-5m: num 10.34 8.64 1.25 1.12 2.13 ...
+```
+
+We can use `ggplot()` to plot this data with a specific `geom_` function called `geom_raster()`. We will make the colour scale in our plot colour-blindness friendly with `scale_fill_viridis_c`, introduced in an [earlier lesson](../episodes/04-intro-to-visualisation.Rmd). We will also use the `coord_quickmap()` function to use an approximate Mercator projection for our plots. This approximation is suitable for small areas that are not too close to the poles. Other coordinate systems are available in `ggplot2` if needed, you can learn about them at their help page `?coord_map`.
+
+
+```r
+ggplot() +
+ geom_raster(data = DSM_TUD_df , aes(x = x, y = y, fill = `tud-dsm-5m`)) +
+ scale_fill_viridis_c(option = "H") + # `option = "H"` provides a contrasting colour scale
+ coord_quickmap()
+```
+
+ +
+Raster plot with `ggplot2` using the viridis color scale
+ +
+Notice that a message is displayed when R creates the histogram:
+
+```
+`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
+```
+
+This message is caused by a default setting in `geom_histogram()` enforcing that there are 30 bins for the data. We can define the number of bins we want in the histogram by giving another value to the `bins` argument. 60 bins, for instance, will give a more detailed histogram of the same distribution:
+
+
+```r
+ggplot() +
+ geom_histogram(data = DSM_TUD_df, aes(`tud-dsm-5m`), bins = 60)
+```
+
+
+
+Notice that a message is displayed when R creates the histogram:
+
+```
+`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
+```
+
+This message is caused by a default setting in `geom_histogram()` enforcing that there are 30 bins for the data. We can define the number of bins we want in the histogram by giving another value to the `bins` argument. 60 bins, for instance, will give a more detailed histogram of the same distribution:
+
+
+```r
+ggplot() +
+ geom_histogram(data = DSM_TUD_df, aes(`tud-dsm-5m`), bins = 60)
+```
+
+ +
+Note that the shape of this histogram looks similar to the previous one that was created using the default of 30 bins. The distribution of elevation values for our Digital Surface Model (DSM) looks reasonable. It is likely that there are no bad data values in this particular raster.
+
+::: challenge
+
+# Challenge: Explore raster metadata
+
+Use `describe()` to determine the following about the `tud-dsm-hill.tif` file:
+
+1. Does this file have the same CRS as `DSM_TUD`?
+2. What is the resolution of the raster data?
+3. How large would a 5x5 pixel area be on the Earth’s surface?
+4. Is the file a multi- or single-band raster?
+
+Note that this file is a hillshade raster. We will learn about hillshades in the [Working with Multi-band Rasters in R](../episodes/17-work-with-multi-band-rasters.Rmd) episode.
+
+::: solution
+
+
+```r
+describe("data/tud-dsm-5m-hill.tif")
+```
+
+```{.output}
+ [1] "Driver: GTiff/GeoTIFF"
+ [2] "Files: data/tud-dsm-5m-hill.tif"
+ [3] "Size is 722, 386"
+ [4] "Coordinate System is:"
+ [5] "PROJCRS[\"Amersfoort / RD New\","
+ [6] " BASEGEOGCRS[\"Amersfoort\","
+ [7] " DATUM[\"Amersfoort\","
+ [8] " ELLIPSOID[\"Bessel 1841\",6377397.155,299.1528128,"
+ [9] " LENGTHUNIT[\"metre\",1]]],"
+[10] " PRIMEM[\"Greenwich\",0,"
+[11] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+[12] " ID[\"EPSG\",4289]],"
+[13] " CONVERSION[\"RD New\","
+[14] " METHOD[\"Oblique Stereographic\","
+[15] " ID[\"EPSG\",9809]],"
+[16] " PARAMETER[\"Latitude of natural origin\",52.1561605555556,"
+[17] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[18] " ID[\"EPSG\",8801]],"
+[19] " PARAMETER[\"Longitude of natural origin\",5.38763888888889,"
+[20] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[21] " ID[\"EPSG\",8802]],"
+[22] " PARAMETER[\"Scale factor at natural origin\",0.9999079,"
+[23] " SCALEUNIT[\"unity\",1],"
+[24] " ID[\"EPSG\",8805]],"
+[25] " PARAMETER[\"False easting\",155000,"
+[26] " LENGTHUNIT[\"metre\",1],"
+[27] " ID[\"EPSG\",8806]],"
+[28] " PARAMETER[\"False northing\",463000,"
+[29] " LENGTHUNIT[\"metre\",1],"
+[30] " ID[\"EPSG\",8807]]],"
+[31] " CS[Cartesian,2],"
+[32] " AXIS[\"easting (X)\",east,"
+[33] " ORDER[1],"
+[34] " LENGTHUNIT[\"metre\",1]],"
+[35] " AXIS[\"northing (Y)\",north,"
+[36] " ORDER[2],"
+[37] " LENGTHUNIT[\"metre\",1]],"
+[38] " USAGE["
+[39] " SCOPE[\"Engineering survey, topographic mapping.\"],"
+[40] " AREA[\"Netherlands - onshore, including Waddenzee, Dutch Wadden Islands and 12-mile offshore coastal zone.\"],"
+[41] " BBOX[50.75,3.2,53.7,7.22]],"
+[42] " ID[\"EPSG\",28992]]"
+[43] "Data axis to CRS axis mapping: 1,2"
+[44] "Origin = (83565.000000000000000,447180.000000000000000)"
+[45] "Pixel Size = (5.000000000000000,-5.000000000000000)"
+[46] "Metadata:"
+[47] " AREA_OR_POINT=Area"
+[48] "Image Structure Metadata:"
+[49] " INTERLEAVE=BAND"
+[50] "Corner Coordinates:"
+[51] "Upper Left ( 83565.000, 447180.000) ( 4d20'49.32\"E, 52d 0'33.67\"N)"
+[52] "Lower Left ( 83565.000, 445250.000) ( 4d20'50.77\"E, 51d59'31.22\"N)"
+[53] "Upper Right ( 87175.000, 447180.000) ( 4d23'58.60\"E, 52d 0'35.30\"N)"
+[54] "Lower Right ( 87175.000, 445250.000) ( 4d23'59.98\"E, 51d59'32.85\"N)"
+[55] "Center ( 85370.000, 446215.000) ( 4d22'24.67\"E, 52d 0' 3.27\"N)"
+[56] "Band 1 Block=722x11 Type=Byte, ColorInterp=Gray"
+[57] " NoData Value=0"
+```
+
+:::
+
+:::
+
+::: callout
+# More resources
+
+- See the manual and tutorials of the `terra` package on [https://rspatial.org/](https://rspatial.org/).
+:::
+
+::::::::::::::::::::::::::::::::::::: keypoints
+
+- The GeoTIFF file format includes metadata about the raster data.
+- To plot raster data with the `ggplot2` package, we need to convert them to data frames.
+- R stores CRS information in the PROJ.4 format.
+- Histograms are useful to identify missing or bad data values.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
diff --git a/14-plot-raster-data.md b/14-plot-raster-data.md
new file mode 100644
index 00000000..606c791c
--- /dev/null
+++ b/14-plot-raster-data.md
@@ -0,0 +1,232 @@
+---
+title: 'Plot Raster Data'
+teaching: 25
+exercises: 10
+---
+
+
+
+:::::::::::::::::::::::::::::::::::::: questions
+
+- How can I create categorized or customized maps of raster data?
+- How can I customize the colour scheme of a raster image?
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::::::::::::::::::::::::::::::::::::: objectives
+
+After completing this episode, participants should be able to…
+
+- Build customized plots for a single band raster using the `ggplot2` package.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::: prereq
+
+# Things you'll need to complete this episode
+
+See the [setup instructions](../learners/setup.md) for detailed information about the software, data, and other prerequisites you will need to work through the examples in this episode.
+
+
+
+:::
+
+In this part, we will plot our raster object using `ggplot2` with customized coloring schemes. We will continue working with the Digital Surface Model (DSM) raster from the [previous episode](../episodes/13-intro-to-raster-data.Rmd).
+
+### Plotting Data Using Breaks
+In the previous plot, our DSM was coloured with a continuous colour range. For clarity and visibility, we may prefer to view the data “symbolized” or coloured according to ranges of values. This is comparable to a “classified” map. For that, we need to tell `ggplot()` how many groups to break our data into and where those breaks should be. To make these decisions, it is useful to first explore the distribution of the data using a bar plot. To begin with, we will use `dplyr`’s `mutate()` function combined with `cut()` to split the data into 3 bins.
+
+
+```r
+DSM_TUD_df <- DSM_TUD_df %>%
+ mutate(fct_elevation = cut(`tud-dsm-5m`, breaks = 3))
+
+ggplot() +
+ geom_bar(data = DSM_TUD_df, aes(fct_elevation))
+```
+
+
+
+Note that the shape of this histogram looks similar to the previous one that was created using the default of 30 bins. The distribution of elevation values for our Digital Surface Model (DSM) looks reasonable. It is likely that there are no bad data values in this particular raster.
+
+::: challenge
+
+# Challenge: Explore raster metadata
+
+Use `describe()` to determine the following about the `tud-dsm-hill.tif` file:
+
+1. Does this file have the same CRS as `DSM_TUD`?
+2. What is the resolution of the raster data?
+3. How large would a 5x5 pixel area be on the Earth’s surface?
+4. Is the file a multi- or single-band raster?
+
+Note that this file is a hillshade raster. We will learn about hillshades in the [Working with Multi-band Rasters in R](../episodes/17-work-with-multi-band-rasters.Rmd) episode.
+
+::: solution
+
+
+```r
+describe("data/tud-dsm-5m-hill.tif")
+```
+
+```{.output}
+ [1] "Driver: GTiff/GeoTIFF"
+ [2] "Files: data/tud-dsm-5m-hill.tif"
+ [3] "Size is 722, 386"
+ [4] "Coordinate System is:"
+ [5] "PROJCRS[\"Amersfoort / RD New\","
+ [6] " BASEGEOGCRS[\"Amersfoort\","
+ [7] " DATUM[\"Amersfoort\","
+ [8] " ELLIPSOID[\"Bessel 1841\",6377397.155,299.1528128,"
+ [9] " LENGTHUNIT[\"metre\",1]]],"
+[10] " PRIMEM[\"Greenwich\",0,"
+[11] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+[12] " ID[\"EPSG\",4289]],"
+[13] " CONVERSION[\"RD New\","
+[14] " METHOD[\"Oblique Stereographic\","
+[15] " ID[\"EPSG\",9809]],"
+[16] " PARAMETER[\"Latitude of natural origin\",52.1561605555556,"
+[17] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[18] " ID[\"EPSG\",8801]],"
+[19] " PARAMETER[\"Longitude of natural origin\",5.38763888888889,"
+[20] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[21] " ID[\"EPSG\",8802]],"
+[22] " PARAMETER[\"Scale factor at natural origin\",0.9999079,"
+[23] " SCALEUNIT[\"unity\",1],"
+[24] " ID[\"EPSG\",8805]],"
+[25] " PARAMETER[\"False easting\",155000,"
+[26] " LENGTHUNIT[\"metre\",1],"
+[27] " ID[\"EPSG\",8806]],"
+[28] " PARAMETER[\"False northing\",463000,"
+[29] " LENGTHUNIT[\"metre\",1],"
+[30] " ID[\"EPSG\",8807]]],"
+[31] " CS[Cartesian,2],"
+[32] " AXIS[\"easting (X)\",east,"
+[33] " ORDER[1],"
+[34] " LENGTHUNIT[\"metre\",1]],"
+[35] " AXIS[\"northing (Y)\",north,"
+[36] " ORDER[2],"
+[37] " LENGTHUNIT[\"metre\",1]],"
+[38] " USAGE["
+[39] " SCOPE[\"Engineering survey, topographic mapping.\"],"
+[40] " AREA[\"Netherlands - onshore, including Waddenzee, Dutch Wadden Islands and 12-mile offshore coastal zone.\"],"
+[41] " BBOX[50.75,3.2,53.7,7.22]],"
+[42] " ID[\"EPSG\",28992]]"
+[43] "Data axis to CRS axis mapping: 1,2"
+[44] "Origin = (83565.000000000000000,447180.000000000000000)"
+[45] "Pixel Size = (5.000000000000000,-5.000000000000000)"
+[46] "Metadata:"
+[47] " AREA_OR_POINT=Area"
+[48] "Image Structure Metadata:"
+[49] " INTERLEAVE=BAND"
+[50] "Corner Coordinates:"
+[51] "Upper Left ( 83565.000, 447180.000) ( 4d20'49.32\"E, 52d 0'33.67\"N)"
+[52] "Lower Left ( 83565.000, 445250.000) ( 4d20'50.77\"E, 51d59'31.22\"N)"
+[53] "Upper Right ( 87175.000, 447180.000) ( 4d23'58.60\"E, 52d 0'35.30\"N)"
+[54] "Lower Right ( 87175.000, 445250.000) ( 4d23'59.98\"E, 51d59'32.85\"N)"
+[55] "Center ( 85370.000, 446215.000) ( 4d22'24.67\"E, 52d 0' 3.27\"N)"
+[56] "Band 1 Block=722x11 Type=Byte, ColorInterp=Gray"
+[57] " NoData Value=0"
+```
+
+:::
+
+:::
+
+::: callout
+# More resources
+
+- See the manual and tutorials of the `terra` package on [https://rspatial.org/](https://rspatial.org/).
+:::
+
+::::::::::::::::::::::::::::::::::::: keypoints
+
+- The GeoTIFF file format includes metadata about the raster data.
+- To plot raster data with the `ggplot2` package, we need to convert them to data frames.
+- R stores CRS information in the PROJ.4 format.
+- Histograms are useful to identify missing or bad data values.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
diff --git a/14-plot-raster-data.md b/14-plot-raster-data.md
new file mode 100644
index 00000000..606c791c
--- /dev/null
+++ b/14-plot-raster-data.md
@@ -0,0 +1,232 @@
+---
+title: 'Plot Raster Data'
+teaching: 25
+exercises: 10
+---
+
+
+
+:::::::::::::::::::::::::::::::::::::: questions
+
+- How can I create categorized or customized maps of raster data?
+- How can I customize the colour scheme of a raster image?
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::::::::::::::::::::::::::::::::::::: objectives
+
+After completing this episode, participants should be able to…
+
+- Build customized plots for a single band raster using the `ggplot2` package.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::: prereq
+
+# Things you'll need to complete this episode
+
+See the [setup instructions](../learners/setup.md) for detailed information about the software, data, and other prerequisites you will need to work through the examples in this episode.
+
+
+
+:::
+
+In this part, we will plot our raster object using `ggplot2` with customized coloring schemes. We will continue working with the Digital Surface Model (DSM) raster from the [previous episode](../episodes/13-intro-to-raster-data.Rmd).
+
+### Plotting Data Using Breaks
+In the previous plot, our DSM was coloured with a continuous colour range. For clarity and visibility, we may prefer to view the data “symbolized” or coloured according to ranges of values. This is comparable to a “classified” map. For that, we need to tell `ggplot()` how many groups to break our data into and where those breaks should be. To make these decisions, it is useful to first explore the distribution of the data using a bar plot. To begin with, we will use `dplyr`’s `mutate()` function combined with `cut()` to split the data into 3 bins.
+
+
+```r
+DSM_TUD_df <- DSM_TUD_df %>%
+ mutate(fct_elevation = cut(`tud-dsm-5m`, breaks = 3))
+
+ggplot() +
+ geom_bar(data = DSM_TUD_df, aes(fct_elevation))
+```
+
+ +
+To see the cut-off values for the groups, we can ask for the levels of `fct_elevation`:
+
+```r
+levels(DSM_TUD_df$fct_elevation)
+```
+
+```{.output}
+[1] "(-5.49,27.1]" "(27.1,59.6]" "(59.6,92.2]"
+```
+
+And we can get the count of values (that is, number of pixels) in each group using `dplyr`’s `count()` function:
+
+```r
+DSM_TUD_df %>%
+ count(fct_elevation)
+```
+
+```{.output}
+ fct_elevation n
+1 (-5.49,27.1] 277100
+2 (27.1,59.6] 1469
+3 (59.6,92.2] 123
+```
+
+We might prefer to customize the cut-off values for these groups. Lets round the cut-off values so that we have groups for the ranges of -10 - 0m, 0 - 5m, and 5 - 100m. To implement this we will give `cut()` a numeric vector of break points instead of the number of breaks we want.
+
+
+```r
+custom_bins <- c(-10, 0, 5, 100)
+
+DSM_TUD_df <- DSM_TUD_df %>%
+ mutate(fct_elevation_cb = cut(`tud-dsm-5m`, breaks = custom_bins))
+
+levels(DSM_TUD_df$fct_elevation_cb)
+```
+
+```{.output}
+[1] "(-10,0]" "(0,5]" "(5,100]"
+```
+
+::: callout
+
+# Data tip
+
+Note that 4 break values will result in 3 bins of data.
+
+The bin intervals are shown using `(` to mean exclusive and `]` to mean inclusive. For example: (0, 10] means “from 0 through 10”.
+
+:::
+
+And now we can plot our bar plot again, using the new groups:
+
+```r
+ggplot() +
+ geom_bar(data = DSM_TUD_df, aes(fct_elevation_cb))
+```
+
+
+
+To see the cut-off values for the groups, we can ask for the levels of `fct_elevation`:
+
+```r
+levels(DSM_TUD_df$fct_elevation)
+```
+
+```{.output}
+[1] "(-5.49,27.1]" "(27.1,59.6]" "(59.6,92.2]"
+```
+
+And we can get the count of values (that is, number of pixels) in each group using `dplyr`’s `count()` function:
+
+```r
+DSM_TUD_df %>%
+ count(fct_elevation)
+```
+
+```{.output}
+ fct_elevation n
+1 (-5.49,27.1] 277100
+2 (27.1,59.6] 1469
+3 (59.6,92.2] 123
+```
+
+We might prefer to customize the cut-off values for these groups. Lets round the cut-off values so that we have groups for the ranges of -10 - 0m, 0 - 5m, and 5 - 100m. To implement this we will give `cut()` a numeric vector of break points instead of the number of breaks we want.
+
+
+```r
+custom_bins <- c(-10, 0, 5, 100)
+
+DSM_TUD_df <- DSM_TUD_df %>%
+ mutate(fct_elevation_cb = cut(`tud-dsm-5m`, breaks = custom_bins))
+
+levels(DSM_TUD_df$fct_elevation_cb)
+```
+
+```{.output}
+[1] "(-10,0]" "(0,5]" "(5,100]"
+```
+
+::: callout
+
+# Data tip
+
+Note that 4 break values will result in 3 bins of data.
+
+The bin intervals are shown using `(` to mean exclusive and `]` to mean inclusive. For example: (0, 10] means “from 0 through 10”.
+
+:::
+
+And now we can plot our bar plot again, using the new groups:
+
+```r
+ggplot() +
+ geom_bar(data = DSM_TUD_df, aes(fct_elevation_cb))
+```
+
+ +
+And we can get the count of values in each group in the same way we did before:
+
+```r
+DSM_TUD_df %>%
+ count(fct_elevation_cb)
+```
+
+```{.output}
+ fct_elevation_cb n
+1 (-10,0] 113877
+2 (0,5] 101446
+3 (5,100] 63369
+```
+
+We can use those groups to plot our raster data, with each group being a different colour:
+
+```r
+ggplot() +
+ geom_raster(data = DSM_TUD_df , aes(x = x, y = y, fill = fct_elevation_cb)) +
+ coord_quickmap()
+```
+
+
+
+And we can get the count of values in each group in the same way we did before:
+
+```r
+DSM_TUD_df %>%
+ count(fct_elevation_cb)
+```
+
+```{.output}
+ fct_elevation_cb n
+1 (-10,0] 113877
+2 (0,5] 101446
+3 (5,100] 63369
+```
+
+We can use those groups to plot our raster data, with each group being a different colour:
+
+```r
+ggplot() +
+ geom_raster(data = DSM_TUD_df , aes(x = x, y = y, fill = fct_elevation_cb)) +
+ coord_quickmap()
+```
+
+ +The plot above uses the default colours inside `ggplot2` for raster objects. We can specify our own colours to make the plot look a little nicer. R has a built in set of colours for plotting terrain, which are built in to the `terrain.colors()` function. Since we have three bins, we want to create a 3-colour palette:
+
+
+```r
+terrain.colors(3)
+```
+
+```{.output}
+[1] "#00A600" "#ECB176" "#F2F2F2"
+```
+
+The `terrain.colors()` function returns hex colours - each of these character strings represents a colour. To use these in our map, we pass them across using the `scale_fill_manual()` function.
+
+```r
+ggplot() +
+ geom_raster(data = DSM_TUD_df , aes(x = x, y = y, fill = fct_elevation_cb)) +
+ scale_fill_manual(values = terrain.colors(3)) +
+ coord_quickmap()
+```
+
+
+The plot above uses the default colours inside `ggplot2` for raster objects. We can specify our own colours to make the plot look a little nicer. R has a built in set of colours for plotting terrain, which are built in to the `terrain.colors()` function. Since we have three bins, we want to create a 3-colour palette:
+
+
+```r
+terrain.colors(3)
+```
+
+```{.output}
+[1] "#00A600" "#ECB176" "#F2F2F2"
+```
+
+The `terrain.colors()` function returns hex colours - each of these character strings represents a colour. To use these in our map, we pass them across using the `scale_fill_manual()` function.
+
+```r
+ggplot() +
+ geom_raster(data = DSM_TUD_df , aes(x = x, y = y, fill = fct_elevation_cb)) +
+ scale_fill_manual(values = terrain.colors(3)) +
+ coord_quickmap()
+```
+
+ +
+## More Plot Formatting
+
+If we need to create multiple plots using the same colour palette, we can create an R object (`my_col`) for the set of colours that we want to use. We can then quickly change the palette across all plots by modifying the `my_col` object, rather than each individual plot.
+
+We can give the legend a more meaningful title by passing a value to the `name` argument of the `scale_fill_manual()` function.
+
+```r
+my_col <- terrain.colors(3)
+
+ggplot() +
+ geom_raster(data = DSM_TUD_df , aes(x = x, y = y,
+ fill = fct_elevation_cb)) +
+ scale_fill_manual(values = my_col, name = "Elevation") +
+ coord_quickmap()
+```
+
+
+
+## More Plot Formatting
+
+If we need to create multiple plots using the same colour palette, we can create an R object (`my_col`) for the set of colours that we want to use. We can then quickly change the palette across all plots by modifying the `my_col` object, rather than each individual plot.
+
+We can give the legend a more meaningful title by passing a value to the `name` argument of the `scale_fill_manual()` function.
+
+```r
+my_col <- terrain.colors(3)
+
+ggplot() +
+ geom_raster(data = DSM_TUD_df , aes(x = x, y = y,
+ fill = fct_elevation_cb)) +
+ scale_fill_manual(values = my_col, name = "Elevation") +
+ coord_quickmap()
+```
+
+ +The axis labels x and y are not necessary, so we can turn them off by passing `element_blank()` to the relevant part of the `theme()` function.
+
+```r
+ggplot() +
+ geom_raster(data = DSM_TUD_df , aes(x = x, y = y,
+ fill = fct_elevation_cb)) +
+ scale_fill_manual(values = my_col, name = "Elevation") +
+ theme(axis.title = element_blank()) +
+ coord_quickmap()
+```
+
+
+The axis labels x and y are not necessary, so we can turn them off by passing `element_blank()` to the relevant part of the `theme()` function.
+
+```r
+ggplot() +
+ geom_raster(data = DSM_TUD_df , aes(x = x, y = y,
+ fill = fct_elevation_cb)) +
+ scale_fill_manual(values = my_col, name = "Elevation") +
+ theme(axis.title = element_blank()) +
+ coord_quickmap()
+```
+
+ +
+
+::: challenge
+
+# Challenge: Plot Using Custom Breaks
+
+Create a plot of the TU Delft Digital Surface Model (`DSM_TUD`) that has:
+
+1. Six classified ranges of values (break points) that are evenly divided among the range of pixel values.
+3. A plot title.
+
+::: solution
+
+
+```r
+DSM_TUD_df <- DSM_TUD_df %>%
+ mutate(fct_elevation_6 = cut(`tud-dsm-5m`, breaks = 6))
+
+levels(DSM_TUD_df$fct_elevation_6)
+```
+
+```{.output}
+[1] "(-5.49,10.9]" "(10.9,27.1]" "(27.1,43.3]" "(43.3,59.6]" "(59.6,75.8]"
+[6] "(75.8,92.2]"
+```
+
+```r
+my_col <- terrain.colors(6)
+
+ggplot() +
+ geom_raster(data = DSM_TUD_df, aes(x = x, y = y,
+ fill = fct_elevation_6)) +
+ scale_fill_manual(values = my_col, name = "Elevation") +
+ coord_quickmap() +
+ labs(title = "Elevation Classes of the Digital Surface Model (DSM)")
+```
+
+
+
+
+::: challenge
+
+# Challenge: Plot Using Custom Breaks
+
+Create a plot of the TU Delft Digital Surface Model (`DSM_TUD`) that has:
+
+1. Six classified ranges of values (break points) that are evenly divided among the range of pixel values.
+3. A plot title.
+
+::: solution
+
+
+```r
+DSM_TUD_df <- DSM_TUD_df %>%
+ mutate(fct_elevation_6 = cut(`tud-dsm-5m`, breaks = 6))
+
+levels(DSM_TUD_df$fct_elevation_6)
+```
+
+```{.output}
+[1] "(-5.49,10.9]" "(10.9,27.1]" "(27.1,43.3]" "(43.3,59.6]" "(59.6,75.8]"
+[6] "(75.8,92.2]"
+```
+
+```r
+my_col <- terrain.colors(6)
+
+ggplot() +
+ geom_raster(data = DSM_TUD_df, aes(x = x, y = y,
+ fill = fct_elevation_6)) +
+ scale_fill_manual(values = my_col, name = "Elevation") +
+ coord_quickmap() +
+ labs(title = "Elevation Classes of the Digital Surface Model (DSM)")
+```
+
+ +
+:::
+
+:::
+
+::::::::::::::::::::::::::::::::::::: keypoints
+
+- Continuous data ranges can be grouped into categories using `mutate()` and `cut()`.
+- Use the built-in `terrain.colors()` or set your preferred colour scheme manually.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
diff --git a/15-reproject-raster-data.md b/15-reproject-raster-data.md
new file mode 100644
index 00000000..5c14c068
--- /dev/null
+++ b/15-reproject-raster-data.md
@@ -0,0 +1,391 @@
+---
+title: 'Reproject Raster Data'
+teaching: 30
+exercises: 2
+---
+
+
+
+:::::::::::::::::::::::::::::::::::::: questions
+
+- How do I work with raster data sets that are in different projections?
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::::::::::::::::::::::::::::::::::::: objectives
+
+After completing this episode, participants should be able to…
+
+- Reproject a raster in R.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::: prereq
+
+# Things you'll need to complete this episode
+
+See the [setup instructions](../learners/setup.md) for detailed information about the software, data, and other prerequisites you will need to work through the examples in this episode.
+
+
+
+:::
+
+Sometimes we encounter raster datasets that do not “line up” when plotted or analysed. Rasters that don’t line up are most often in different Coordinate Reference Systems (CRS). This episode explains how to deal with rasters in different CRS. It will walk through reprojecting rasters in R using the `project()` function in the `terra` package.
+
+
+
+## Raster Projection
+
+For this episode, we will be working with the Digital Terrain Model data. This differs from the surface model data we’ve been working with so far in that the digital surface model (DSM) includes the tops of trees and buildings, while the digital terrain model (DTM) shows the ground level.
+
+We’ll be looking at another model (the canopy/building height model, or CHM) in [a later episode](../episodes/16-raster-calculations.Rmd) and will see how to calculate the CHM from the DSM and DTM. Here, we will create a map of the Digital Terrain Model (`DTM_TUD`) of TU Delft and its surrounding draped (or layered) on top of the hillshade (`DTM_hill_TUD`).
+
+:::::::::::::::::::::::::::::::::::::: callout
+
+### Layering Rasters: Hillshade
+
+We can layer a raster on top of a hillshade raster for the same area, and use a transparency factor to create a 3-dimensional shaded effect. A hillshade is a raster that maps the terrain using light and shadow to create a 3D-looking image that you would see from above when viewing the terrain. We will add a custom colour, making the plot grey.
+
+Read more about layering raster [here](https://datacarpentry.org/r-raster-vector-geospatial/instructor/02-raster-plot.html#layering-rasters).
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+First, we need to import the DTM and DTM hillshade data.
+
+
+```r
+DTM_TUD <- rast("data/tud-dtm-5m.tif")
+DTM_hill_TUD <- rast("data/tud-dtm-5m-hill-WGS84.tif")
+```
+
+Next, we will convert each of these datasets to a data frame for plotting with `ggplot`.
+
+```r
+DTM_TUD_df <- as.data.frame(DTM_TUD, xy = TRUE)
+DTM_hill_TUD_df <- as.data.frame(DTM_hill_TUD, xy = TRUE)
+```
+
+Now we can create a map of the DTM layered over the hillshade.
+
+```r
+ggplot() +
+ geom_raster(data = DTM_TUD_df ,
+ aes(x = x, y = y,
+ fill = `tud-dtm-5m`)) +
+ geom_raster(data = DTM_hill_TUD_df,
+ aes(x = x, y = y,
+ alpha = `tud-dtm-5m-hill`)) +
+ scale_fill_gradientn(name = "Elevation", colors = terrain.colors(10)) +
+ coord_quickmap()
+```
+
+
+
+:::
+
+:::
+
+::::::::::::::::::::::::::::::::::::: keypoints
+
+- Continuous data ranges can be grouped into categories using `mutate()` and `cut()`.
+- Use the built-in `terrain.colors()` or set your preferred colour scheme manually.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
diff --git a/15-reproject-raster-data.md b/15-reproject-raster-data.md
new file mode 100644
index 00000000..5c14c068
--- /dev/null
+++ b/15-reproject-raster-data.md
@@ -0,0 +1,391 @@
+---
+title: 'Reproject Raster Data'
+teaching: 30
+exercises: 2
+---
+
+
+
+:::::::::::::::::::::::::::::::::::::: questions
+
+- How do I work with raster data sets that are in different projections?
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::::::::::::::::::::::::::::::::::::: objectives
+
+After completing this episode, participants should be able to…
+
+- Reproject a raster in R.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::: prereq
+
+# Things you'll need to complete this episode
+
+See the [setup instructions](../learners/setup.md) for detailed information about the software, data, and other prerequisites you will need to work through the examples in this episode.
+
+
+
+:::
+
+Sometimes we encounter raster datasets that do not “line up” when plotted or analysed. Rasters that don’t line up are most often in different Coordinate Reference Systems (CRS). This episode explains how to deal with rasters in different CRS. It will walk through reprojecting rasters in R using the `project()` function in the `terra` package.
+
+
+
+## Raster Projection
+
+For this episode, we will be working with the Digital Terrain Model data. This differs from the surface model data we’ve been working with so far in that the digital surface model (DSM) includes the tops of trees and buildings, while the digital terrain model (DTM) shows the ground level.
+
+We’ll be looking at another model (the canopy/building height model, or CHM) in [a later episode](../episodes/16-raster-calculations.Rmd) and will see how to calculate the CHM from the DSM and DTM. Here, we will create a map of the Digital Terrain Model (`DTM_TUD`) of TU Delft and its surrounding draped (or layered) on top of the hillshade (`DTM_hill_TUD`).
+
+:::::::::::::::::::::::::::::::::::::: callout
+
+### Layering Rasters: Hillshade
+
+We can layer a raster on top of a hillshade raster for the same area, and use a transparency factor to create a 3-dimensional shaded effect. A hillshade is a raster that maps the terrain using light and shadow to create a 3D-looking image that you would see from above when viewing the terrain. We will add a custom colour, making the plot grey.
+
+Read more about layering raster [here](https://datacarpentry.org/r-raster-vector-geospatial/instructor/02-raster-plot.html#layering-rasters).
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+First, we need to import the DTM and DTM hillshade data.
+
+
+```r
+DTM_TUD <- rast("data/tud-dtm-5m.tif")
+DTM_hill_TUD <- rast("data/tud-dtm-5m-hill-WGS84.tif")
+```
+
+Next, we will convert each of these datasets to a data frame for plotting with `ggplot`.
+
+```r
+DTM_TUD_df <- as.data.frame(DTM_TUD, xy = TRUE)
+DTM_hill_TUD_df <- as.data.frame(DTM_hill_TUD, xy = TRUE)
+```
+
+Now we can create a map of the DTM layered over the hillshade.
+
+```r
+ggplot() +
+ geom_raster(data = DTM_TUD_df ,
+ aes(x = x, y = y,
+ fill = `tud-dtm-5m`)) +
+ geom_raster(data = DTM_hill_TUD_df,
+ aes(x = x, y = y,
+ alpha = `tud-dtm-5m-hill`)) +
+ scale_fill_gradientn(name = "Elevation", colors = terrain.colors(10)) +
+ coord_quickmap()
+```
+
+ +Our results are curious - neither the Digital Terrain Model (`DTM_TUD_df`) nor the DTM Hillshade (`DTM_hill_TUD_df`) plotted. Let’s try to plot the DTM on its own to make sure there are data there.
+
+
+```r
+ggplot() +
+ geom_raster(data = DTM_TUD_df,
+ aes(x = x, y = y,
+ fill = `tud-dtm-5m`)) +
+ scale_fill_gradientn(name = "Elevation", colors = terrain.colors(10)) +
+ coord_quickmap()
+```
+
+
+Our results are curious - neither the Digital Terrain Model (`DTM_TUD_df`) nor the DTM Hillshade (`DTM_hill_TUD_df`) plotted. Let’s try to plot the DTM on its own to make sure there are data there.
+
+
+```r
+ggplot() +
+ geom_raster(data = DTM_TUD_df,
+ aes(x = x, y = y,
+ fill = `tud-dtm-5m`)) +
+ scale_fill_gradientn(name = "Elevation", colors = terrain.colors(10)) +
+ coord_quickmap()
+```
+
+ +
+Our DTM seems to contain data and plots just fine.
+
+Next we plot the DTM Hillshade on its own to see whether everything is OK.
+
+
+```r
+ggplot() +
+ geom_raster(data = DTM_hill_TUD_df,
+ aes(x = x, y = y,
+ alpha = `tud-dtm-5m-hill`)) +
+ coord_quickmap()
+```
+
+
+
+Our DTM seems to contain data and plots just fine.
+
+Next we plot the DTM Hillshade on its own to see whether everything is OK.
+
+
+```r
+ggplot() +
+ geom_raster(data = DTM_hill_TUD_df,
+ aes(x = x, y = y,
+ alpha = `tud-dtm-5m-hill`)) +
+ coord_quickmap()
+```
+
+ +
+If we look at the axes, we can see that the units, and therefore the projections, of the two rasters are different. When this is the case, `ggplot2` won’t render the image. It won’t even throw an error message to tell you something has gone wrong. We can look at Coordinate Reference Systems (CRSs) of the DTM and the hillshade data to see how they differ.
+
+::: challenge
+
+# Exercise
+
+View the CRS for each of these two datasets. What projection does each use?
+
+::: solution
+
+
+```r
+crs(DTM_TUD, parse = TRUE)
+```
+
+```{.output}
+ [1] "PROJCRS[\"Amersfoort / RD New\","
+ [2] " BASEGEOGCRS[\"Amersfoort\","
+ [3] " DATUM[\"Amersfoort\","
+ [4] " ELLIPSOID[\"Bessel 1841\",6377397.155,299.1528128,"
+ [5] " LENGTHUNIT[\"metre\",1]]],"
+ [6] " PRIMEM[\"Greenwich\",0,"
+ [7] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+ [8] " ID[\"EPSG\",4289]],"
+ [9] " CONVERSION[\"RD New\","
+[10] " METHOD[\"Oblique Stereographic\","
+[11] " ID[\"EPSG\",9809]],"
+[12] " PARAMETER[\"Latitude of natural origin\",52.1561605555556,"
+[13] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[14] " ID[\"EPSG\",8801]],"
+[15] " PARAMETER[\"Longitude of natural origin\",5.38763888888889,"
+[16] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[17] " ID[\"EPSG\",8802]],"
+[18] " PARAMETER[\"Scale factor at natural origin\",0.9999079,"
+[19] " SCALEUNIT[\"unity\",1],"
+[20] " ID[\"EPSG\",8805]],"
+[21] " PARAMETER[\"False easting\",155000,"
+[22] " LENGTHUNIT[\"metre\",1],"
+[23] " ID[\"EPSG\",8806]],"
+[24] " PARAMETER[\"False northing\",463000,"
+[25] " LENGTHUNIT[\"metre\",1],"
+[26] " ID[\"EPSG\",8807]]],"
+[27] " CS[Cartesian,2],"
+[28] " AXIS[\"easting (X)\",east,"
+[29] " ORDER[1],"
+[30] " LENGTHUNIT[\"metre\",1]],"
+[31] " AXIS[\"northing (Y)\",north,"
+[32] " ORDER[2],"
+[33] " LENGTHUNIT[\"metre\",1]],"
+[34] " USAGE["
+[35] " SCOPE[\"Engineering survey, topographic mapping.\"],"
+[36] " AREA[\"Netherlands - onshore, including Waddenzee, Dutch Wadden Islands and 12-mile offshore coastal zone.\"],"
+[37] " BBOX[50.75,3.2,53.7,7.22]],"
+[38] " ID[\"EPSG\",28992]]"
+```
+
+
+```r
+crs(DTM_hill_TUD, parse = TRUE)
+```
+
+```{.output}
+ [1] "GEOGCRS[\"WGS 84\","
+ [2] " DATUM[\"World Geodetic System 1984\","
+ [3] " ELLIPSOID[\"WGS 84\",6378137,298.257223563,"
+ [4] " LENGTHUNIT[\"metre\",1]]],"
+ [5] " PRIMEM[\"Greenwich\",0,"
+ [6] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+ [7] " CS[ellipsoidal,2],"
+ [8] " AXIS[\"geodetic latitude (Lat)\",north,"
+ [9] " ORDER[1],"
+[10] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+[11] " AXIS[\"geodetic longitude (Lon)\",east,"
+[12] " ORDER[2],"
+[13] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+[14] " ID[\"EPSG\",4326]]"
+```
+`DTM_TUD` is in the Amersfoort / RD New projection, whereas `DTM_hill_TUD` is in WGS 84.
+
+:::
+
+:::
+
+Because the two rasters are in different CRS, they don’t line up when plotted in R. We need to reproject (or change the projection of) `DTM_hill_TUD` into the Amersfoort / RD New CRS.
+
+## Reproject Rasters
+
+We can use the `project()` function to reproject a raster into a new CRS. Keep in mind that reprojection only works when you first have a defined CRS for the raster object that you want to reproject. It cannot be used if no CRS is defined. Lucky for us, DTM_hill_TUD has a defined CRS.
+
+::: callout
+
+# Data tip
+
+When we reproject a raster, we move it from one “grid” to another. Thus, we are modifying the data! Keep this in mind as we work with raster data.
+
+:::
+
+To use the `project()` function, we need to define two things:
+
+1. the object we want to reproject and
+2. the CRS that we want to reproject it to.
+
+The syntax is `project(RasterObject, crs)`
+
+We want the CRS of our hillshade to match the `DTM_TUD` raster. We can thus assign the CRS of our `DTM_TUD` to our hillshade within the `project()` function as follows: `crs(DTM_TUD)`. Note that we are using the `project()` function on the raster object, not the `data.frame()` we use for plotting with `ggplot`.
+
+First we will reproject our `DTM_hill_TUD` raster data to match the `DTM_TUD` raster CRS:
+
+```r
+DTM_hill_EPSG28992_TUD <- project(DTM_hill_TUD,
+ crs(DTM_TUD))
+```
+
+Now we can compare the CRS of our original DTM hillshade and our new DTM hillshade, to see how they are different.
+
+```r
+crs(DTM_hill_EPSG28992_TUD, parse = TRUE)
+```
+
+```{.output}
+ [1] "PROJCRS[\"Amersfoort / RD New\","
+ [2] " BASEGEOGCRS[\"Amersfoort\","
+ [3] " DATUM[\"Amersfoort\","
+ [4] " ELLIPSOID[\"Bessel 1841\",6377397.155,299.1528128,"
+ [5] " LENGTHUNIT[\"metre\",1]]],"
+ [6] " PRIMEM[\"Greenwich\",0,"
+ [7] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+ [8] " ID[\"EPSG\",4289]],"
+ [9] " CONVERSION[\"RD New\","
+[10] " METHOD[\"Oblique Stereographic\","
+[11] " ID[\"EPSG\",9809]],"
+[12] " PARAMETER[\"Latitude of natural origin\",52.1561605555556,"
+[13] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[14] " ID[\"EPSG\",8801]],"
+[15] " PARAMETER[\"Longitude of natural origin\",5.38763888888889,"
+[16] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[17] " ID[\"EPSG\",8802]],"
+[18] " PARAMETER[\"Scale factor at natural origin\",0.9999079,"
+[19] " SCALEUNIT[\"unity\",1],"
+[20] " ID[\"EPSG\",8805]],"
+[21] " PARAMETER[\"False easting\",155000,"
+[22] " LENGTHUNIT[\"metre\",1],"
+[23] " ID[\"EPSG\",8806]],"
+[24] " PARAMETER[\"False northing\",463000,"
+[25] " LENGTHUNIT[\"metre\",1],"
+[26] " ID[\"EPSG\",8807]]],"
+[27] " CS[Cartesian,2],"
+[28] " AXIS[\"easting (X)\",east,"
+[29] " ORDER[1],"
+[30] " LENGTHUNIT[\"metre\",1]],"
+[31] " AXIS[\"northing (Y)\",north,"
+[32] " ORDER[2],"
+[33] " LENGTHUNIT[\"metre\",1]],"
+[34] " USAGE["
+[35] " SCOPE[\"Engineering survey, topographic mapping.\"],"
+[36] " AREA[\"Netherlands - onshore, including Waddenzee, Dutch Wadden Islands and 12-mile offshore coastal zone.\"],"
+[37] " BBOX[50.75,3.2,53.7,7.22]],"
+[38] " ID[\"EPSG\",28992]]"
+```
+
+```r
+crs(DTM_hill_TUD, parse = TRUE)
+```
+
+```{.output}
+ [1] "GEOGCRS[\"WGS 84\","
+ [2] " DATUM[\"World Geodetic System 1984\","
+ [3] " ELLIPSOID[\"WGS 84\",6378137,298.257223563,"
+ [4] " LENGTHUNIT[\"metre\",1]]],"
+ [5] " PRIMEM[\"Greenwich\",0,"
+ [6] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+ [7] " CS[ellipsoidal,2],"
+ [8] " AXIS[\"geodetic latitude (Lat)\",north,"
+ [9] " ORDER[1],"
+[10] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+[11] " AXIS[\"geodetic longitude (Lon)\",east,"
+[12] " ORDER[2],"
+[13] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+[14] " ID[\"EPSG\",4326]]"
+```
+
+We can also compare the extent of the two objects.
+
+```r
+ext(DTM_hill_EPSG28992_TUD)
+```
+
+```{.output}
+SpatExtent : 83537.3768729672, 87200.5199626113, 445202.584641046, 447230.395994242 (xmin, xmax, ymin, ymax)
+```
+
+```r
+ext(DTM_hill_TUD)
+```
+
+```{.output}
+SpatExtent : 4.34674898644234, 4.39970436836596, 51.9910492930106, 52.0088368700157 (xmin, xmax, ymin, ymax)
+```
+
+## Deal with Raster Resolution
+
+Let’s next have a look at the resolution of our reprojected hillshade versus our original data.
+
+
+```r
+res(DTM_hill_EPSG28992_TUD)
+```
+
+```{.output}
+[1] 5.03179 5.03179
+```
+
+
+```r
+res(DTM_TUD)
+```
+
+```{.output}
+[1] 5 5
+```
+
+These two resolutions are different, but they’re representing the same data. We can tell R to force our newly reprojected raster to be the same as `DTM_TUD` by adding a line of code `res = res(DTM_TUD)` within the `project()` function.
+
+```r
+DTM_hill_EPSG28992_TUD <- project(DTM_hill_TUD,
+ crs(DTM_TUD),
+ res = res(DTM_TUD))
+```
+
+Now both our resolutions and our CRSs match, so we can plot these two data sets together. Let’s double-check our resolution to be sure:
+
+```r
+res(DTM_hill_EPSG28992_TUD)
+```
+
+```{.output}
+[1] 5 5
+```
+
+```r
+res(DTM_TUD)
+```
+
+```{.output}
+[1] 5 5
+```
+
+For plotting with `ggplot()`, we will need to create a data frame from our newly reprojected raster.
+
+```r
+DTM_hill_TUD_2_df <- as.data.frame(DTM_hill_EPSG28992_TUD, xy = TRUE)
+```
+
+We can now create a plot of this data.
+
+```r
+ggplot() +
+ geom_raster(data = DTM_TUD_df ,
+ aes(x = x, y = y,
+ fill = `tud-dtm-5m`)) +
+ geom_raster(data = DTM_hill_TUD_2_df,
+ aes(x = x, y = y,
+ alpha = `tud-dtm-5m-hill`)) +
+ scale_fill_gradientn(name = "Elevation", colors = terrain.colors(10)) +
+ coord_equal()
+```
+
+
+
+If we look at the axes, we can see that the units, and therefore the projections, of the two rasters are different. When this is the case, `ggplot2` won’t render the image. It won’t even throw an error message to tell you something has gone wrong. We can look at Coordinate Reference Systems (CRSs) of the DTM and the hillshade data to see how they differ.
+
+::: challenge
+
+# Exercise
+
+View the CRS for each of these two datasets. What projection does each use?
+
+::: solution
+
+
+```r
+crs(DTM_TUD, parse = TRUE)
+```
+
+```{.output}
+ [1] "PROJCRS[\"Amersfoort / RD New\","
+ [2] " BASEGEOGCRS[\"Amersfoort\","
+ [3] " DATUM[\"Amersfoort\","
+ [4] " ELLIPSOID[\"Bessel 1841\",6377397.155,299.1528128,"
+ [5] " LENGTHUNIT[\"metre\",1]]],"
+ [6] " PRIMEM[\"Greenwich\",0,"
+ [7] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+ [8] " ID[\"EPSG\",4289]],"
+ [9] " CONVERSION[\"RD New\","
+[10] " METHOD[\"Oblique Stereographic\","
+[11] " ID[\"EPSG\",9809]],"
+[12] " PARAMETER[\"Latitude of natural origin\",52.1561605555556,"
+[13] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[14] " ID[\"EPSG\",8801]],"
+[15] " PARAMETER[\"Longitude of natural origin\",5.38763888888889,"
+[16] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[17] " ID[\"EPSG\",8802]],"
+[18] " PARAMETER[\"Scale factor at natural origin\",0.9999079,"
+[19] " SCALEUNIT[\"unity\",1],"
+[20] " ID[\"EPSG\",8805]],"
+[21] " PARAMETER[\"False easting\",155000,"
+[22] " LENGTHUNIT[\"metre\",1],"
+[23] " ID[\"EPSG\",8806]],"
+[24] " PARAMETER[\"False northing\",463000,"
+[25] " LENGTHUNIT[\"metre\",1],"
+[26] " ID[\"EPSG\",8807]]],"
+[27] " CS[Cartesian,2],"
+[28] " AXIS[\"easting (X)\",east,"
+[29] " ORDER[1],"
+[30] " LENGTHUNIT[\"metre\",1]],"
+[31] " AXIS[\"northing (Y)\",north,"
+[32] " ORDER[2],"
+[33] " LENGTHUNIT[\"metre\",1]],"
+[34] " USAGE["
+[35] " SCOPE[\"Engineering survey, topographic mapping.\"],"
+[36] " AREA[\"Netherlands - onshore, including Waddenzee, Dutch Wadden Islands and 12-mile offshore coastal zone.\"],"
+[37] " BBOX[50.75,3.2,53.7,7.22]],"
+[38] " ID[\"EPSG\",28992]]"
+```
+
+
+```r
+crs(DTM_hill_TUD, parse = TRUE)
+```
+
+```{.output}
+ [1] "GEOGCRS[\"WGS 84\","
+ [2] " DATUM[\"World Geodetic System 1984\","
+ [3] " ELLIPSOID[\"WGS 84\",6378137,298.257223563,"
+ [4] " LENGTHUNIT[\"metre\",1]]],"
+ [5] " PRIMEM[\"Greenwich\",0,"
+ [6] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+ [7] " CS[ellipsoidal,2],"
+ [8] " AXIS[\"geodetic latitude (Lat)\",north,"
+ [9] " ORDER[1],"
+[10] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+[11] " AXIS[\"geodetic longitude (Lon)\",east,"
+[12] " ORDER[2],"
+[13] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+[14] " ID[\"EPSG\",4326]]"
+```
+`DTM_TUD` is in the Amersfoort / RD New projection, whereas `DTM_hill_TUD` is in WGS 84.
+
+:::
+
+:::
+
+Because the two rasters are in different CRS, they don’t line up when plotted in R. We need to reproject (or change the projection of) `DTM_hill_TUD` into the Amersfoort / RD New CRS.
+
+## Reproject Rasters
+
+We can use the `project()` function to reproject a raster into a new CRS. Keep in mind that reprojection only works when you first have a defined CRS for the raster object that you want to reproject. It cannot be used if no CRS is defined. Lucky for us, DTM_hill_TUD has a defined CRS.
+
+::: callout
+
+# Data tip
+
+When we reproject a raster, we move it from one “grid” to another. Thus, we are modifying the data! Keep this in mind as we work with raster data.
+
+:::
+
+To use the `project()` function, we need to define two things:
+
+1. the object we want to reproject and
+2. the CRS that we want to reproject it to.
+
+The syntax is `project(RasterObject, crs)`
+
+We want the CRS of our hillshade to match the `DTM_TUD` raster. We can thus assign the CRS of our `DTM_TUD` to our hillshade within the `project()` function as follows: `crs(DTM_TUD)`. Note that we are using the `project()` function on the raster object, not the `data.frame()` we use for plotting with `ggplot`.
+
+First we will reproject our `DTM_hill_TUD` raster data to match the `DTM_TUD` raster CRS:
+
+```r
+DTM_hill_EPSG28992_TUD <- project(DTM_hill_TUD,
+ crs(DTM_TUD))
+```
+
+Now we can compare the CRS of our original DTM hillshade and our new DTM hillshade, to see how they are different.
+
+```r
+crs(DTM_hill_EPSG28992_TUD, parse = TRUE)
+```
+
+```{.output}
+ [1] "PROJCRS[\"Amersfoort / RD New\","
+ [2] " BASEGEOGCRS[\"Amersfoort\","
+ [3] " DATUM[\"Amersfoort\","
+ [4] " ELLIPSOID[\"Bessel 1841\",6377397.155,299.1528128,"
+ [5] " LENGTHUNIT[\"metre\",1]]],"
+ [6] " PRIMEM[\"Greenwich\",0,"
+ [7] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+ [8] " ID[\"EPSG\",4289]],"
+ [9] " CONVERSION[\"RD New\","
+[10] " METHOD[\"Oblique Stereographic\","
+[11] " ID[\"EPSG\",9809]],"
+[12] " PARAMETER[\"Latitude of natural origin\",52.1561605555556,"
+[13] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[14] " ID[\"EPSG\",8801]],"
+[15] " PARAMETER[\"Longitude of natural origin\",5.38763888888889,"
+[16] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[17] " ID[\"EPSG\",8802]],"
+[18] " PARAMETER[\"Scale factor at natural origin\",0.9999079,"
+[19] " SCALEUNIT[\"unity\",1],"
+[20] " ID[\"EPSG\",8805]],"
+[21] " PARAMETER[\"False easting\",155000,"
+[22] " LENGTHUNIT[\"metre\",1],"
+[23] " ID[\"EPSG\",8806]],"
+[24] " PARAMETER[\"False northing\",463000,"
+[25] " LENGTHUNIT[\"metre\",1],"
+[26] " ID[\"EPSG\",8807]]],"
+[27] " CS[Cartesian,2],"
+[28] " AXIS[\"easting (X)\",east,"
+[29] " ORDER[1],"
+[30] " LENGTHUNIT[\"metre\",1]],"
+[31] " AXIS[\"northing (Y)\",north,"
+[32] " ORDER[2],"
+[33] " LENGTHUNIT[\"metre\",1]],"
+[34] " USAGE["
+[35] " SCOPE[\"Engineering survey, topographic mapping.\"],"
+[36] " AREA[\"Netherlands - onshore, including Waddenzee, Dutch Wadden Islands and 12-mile offshore coastal zone.\"],"
+[37] " BBOX[50.75,3.2,53.7,7.22]],"
+[38] " ID[\"EPSG\",28992]]"
+```
+
+```r
+crs(DTM_hill_TUD, parse = TRUE)
+```
+
+```{.output}
+ [1] "GEOGCRS[\"WGS 84\","
+ [2] " DATUM[\"World Geodetic System 1984\","
+ [3] " ELLIPSOID[\"WGS 84\",6378137,298.257223563,"
+ [4] " LENGTHUNIT[\"metre\",1]]],"
+ [5] " PRIMEM[\"Greenwich\",0,"
+ [6] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+ [7] " CS[ellipsoidal,2],"
+ [8] " AXIS[\"geodetic latitude (Lat)\",north,"
+ [9] " ORDER[1],"
+[10] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+[11] " AXIS[\"geodetic longitude (Lon)\",east,"
+[12] " ORDER[2],"
+[13] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+[14] " ID[\"EPSG\",4326]]"
+```
+
+We can also compare the extent of the two objects.
+
+```r
+ext(DTM_hill_EPSG28992_TUD)
+```
+
+```{.output}
+SpatExtent : 83537.3768729672, 87200.5199626113, 445202.584641046, 447230.395994242 (xmin, xmax, ymin, ymax)
+```
+
+```r
+ext(DTM_hill_TUD)
+```
+
+```{.output}
+SpatExtent : 4.34674898644234, 4.39970436836596, 51.9910492930106, 52.0088368700157 (xmin, xmax, ymin, ymax)
+```
+
+## Deal with Raster Resolution
+
+Let’s next have a look at the resolution of our reprojected hillshade versus our original data.
+
+
+```r
+res(DTM_hill_EPSG28992_TUD)
+```
+
+```{.output}
+[1] 5.03179 5.03179
+```
+
+
+```r
+res(DTM_TUD)
+```
+
+```{.output}
+[1] 5 5
+```
+
+These two resolutions are different, but they’re representing the same data. We can tell R to force our newly reprojected raster to be the same as `DTM_TUD` by adding a line of code `res = res(DTM_TUD)` within the `project()` function.
+
+```r
+DTM_hill_EPSG28992_TUD <- project(DTM_hill_TUD,
+ crs(DTM_TUD),
+ res = res(DTM_TUD))
+```
+
+Now both our resolutions and our CRSs match, so we can plot these two data sets together. Let’s double-check our resolution to be sure:
+
+```r
+res(DTM_hill_EPSG28992_TUD)
+```
+
+```{.output}
+[1] 5 5
+```
+
+```r
+res(DTM_TUD)
+```
+
+```{.output}
+[1] 5 5
+```
+
+For plotting with `ggplot()`, we will need to create a data frame from our newly reprojected raster.
+
+```r
+DTM_hill_TUD_2_df <- as.data.frame(DTM_hill_EPSG28992_TUD, xy = TRUE)
+```
+
+We can now create a plot of this data.
+
+```r
+ggplot() +
+ geom_raster(data = DTM_TUD_df ,
+ aes(x = x, y = y,
+ fill = `tud-dtm-5m`)) +
+ geom_raster(data = DTM_hill_TUD_2_df,
+ aes(x = x, y = y,
+ alpha = `tud-dtm-5m-hill`)) +
+ scale_fill_gradientn(name = "Elevation", colors = terrain.colors(10)) +
+ coord_equal()
+```
+
+ +
+We have now successfully draped the Digital Terrain Model on top of our hillshade to produce a nice looking, textured map!
+
+
+
+::::::::::::::::::::::::::::::::::::: keypoints
+
+- In order to plot two raster data sets together, they must be in the same CRS.
+- Use the `project()` function to convert between CRSs.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
diff --git a/16-raster-calculations.md b/16-raster-calculations.md
new file mode 100644
index 00000000..b2ed9b34
--- /dev/null
+++ b/16-raster-calculations.md
@@ -0,0 +1,324 @@
+---
+title: 'Raster Calculations'
+teaching: 25
+exercises: 5
+---
+
+
+
+:::::::::::::::::::::::::::::::::::::: questions
+
+- How do I subtract one raster from another and extract pixel values for defined locations?
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::::::::::::::::::::::::::::::::::::: objectives
+
+After completing this episode, participants should be able to…
+
+- Perform a subtraction between two rasters using raster math.
+- Export raster data as a GeoTIFF file.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::: prereq
+
+# Things you'll need to complete this episode
+
+See the [setup instructions](../learners/setup.md) for detailed information about the software, data, and other prerequisites you will need to work through the examples in this episode.
+
+
+
+:::
+
+We often want to combine values of and perform calculations on rasters to create a new output raster. This episode covers how to subtract one raster from another using basic raster math.
+
+## Raster calculations in R
+
+We often want to perform calculations on two or more rasters to create a new output raster. For example, if we are interested in mapping the heights of trees and buildings across an entire field site, we might want to calculate the difference between the Digital Surface Model (DSM, tops of trees and buildings) and the Digital Terrain Model (DTM, ground level). The resulting dataset is referred to as a Canopy Height Model (CHM) and represents the actual height of trees, buildings, etc. with the influence of ground elevation removed.
+
+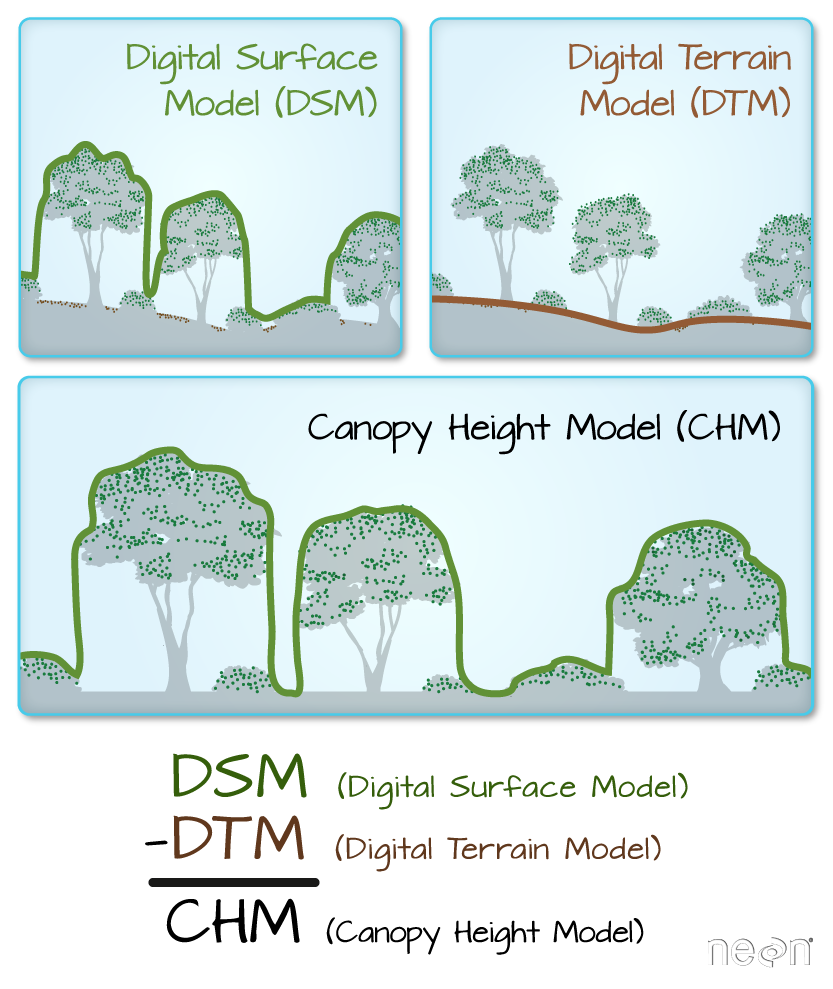
+
+## Load the Data
+
+For this episode, we will use the DTM and DSM data which we already have loaded from previous episodes.
+
+We use the `describe()` function to view information about the DTM and DSM data files.
+
+```r
+describe("data/tud-dtm-5m.tif")
+```
+
+```{.output}
+ [1] "Driver: GTiff/GeoTIFF"
+ [2] "Files: data/tud-dtm-5m.tif"
+ [3] "Size is 722, 386"
+ [4] "Coordinate System is:"
+ [5] "PROJCRS[\"Amersfoort / RD New\","
+ [6] " BASEGEOGCRS[\"Amersfoort\","
+ [7] " DATUM[\"Amersfoort\","
+ [8] " ELLIPSOID[\"Bessel 1841\",6377397.155,299.1528128,"
+ [9] " LENGTHUNIT[\"metre\",1]]],"
+[10] " PRIMEM[\"Greenwich\",0,"
+[11] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+[12] " ID[\"EPSG\",4289]],"
+[13] " CONVERSION[\"RD New\","
+[14] " METHOD[\"Oblique Stereographic\","
+[15] " ID[\"EPSG\",9809]],"
+[16] " PARAMETER[\"Latitude of natural origin\",52.1561605555556,"
+[17] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[18] " ID[\"EPSG\",8801]],"
+[19] " PARAMETER[\"Longitude of natural origin\",5.38763888888889,"
+[20] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[21] " ID[\"EPSG\",8802]],"
+[22] " PARAMETER[\"Scale factor at natural origin\",0.9999079,"
+[23] " SCALEUNIT[\"unity\",1],"
+[24] " ID[\"EPSG\",8805]],"
+[25] " PARAMETER[\"False easting\",155000,"
+[26] " LENGTHUNIT[\"metre\",1],"
+[27] " ID[\"EPSG\",8806]],"
+[28] " PARAMETER[\"False northing\",463000,"
+[29] " LENGTHUNIT[\"metre\",1],"
+[30] " ID[\"EPSG\",8807]]],"
+[31] " CS[Cartesian,2],"
+[32] " AXIS[\"easting (X)\",east,"
+[33] " ORDER[1],"
+[34] " LENGTHUNIT[\"metre\",1]],"
+[35] " AXIS[\"northing (Y)\",north,"
+[36] " ORDER[2],"
+[37] " LENGTHUNIT[\"metre\",1]],"
+[38] " USAGE["
+[39] " SCOPE[\"Engineering survey, topographic mapping.\"],"
+[40] " AREA[\"Netherlands - onshore, including Waddenzee, Dutch Wadden Islands and 12-mile offshore coastal zone.\"],"
+[41] " BBOX[50.75,3.2,53.7,7.22]],"
+[42] " ID[\"EPSG\",28992]]"
+[43] "Data axis to CRS axis mapping: 1,2"
+[44] "Origin = (83565.000000000000000,447180.000000000000000)"
+[45] "Pixel Size = (5.000000000000000,-5.000000000000000)"
+[46] "Metadata:"
+[47] " AREA_OR_POINT=Area"
+[48] "Image Structure Metadata:"
+[49] " INTERLEAVE=BAND"
+[50] "Corner Coordinates:"
+[51] "Upper Left ( 83565.000, 447180.000) ( 4d20'49.32\"E, 52d 0'33.67\"N)"
+[52] "Lower Left ( 83565.000, 445250.000) ( 4d20'50.77\"E, 51d59'31.22\"N)"
+[53] "Upper Right ( 87175.000, 447180.000) ( 4d23'58.60\"E, 52d 0'35.30\"N)"
+[54] "Lower Right ( 87175.000, 445250.000) ( 4d23'59.98\"E, 51d59'32.85\"N)"
+[55] "Center ( 85370.000, 446215.000) ( 4d22'24.67\"E, 52d 0' 3.27\"N)"
+[56] "Band 1 Block=722x2 Type=Float32, ColorInterp=Gray"
+```
+
+```r
+describe("data/tud-dsm-5m.tif")
+```
+
+```{.output}
+ [1] "Driver: GTiff/GeoTIFF"
+ [2] "Files: data/tud-dsm-5m.tif"
+ [3] "Size is 722, 386"
+ [4] "Coordinate System is:"
+ [5] "PROJCRS[\"Amersfoort / RD New\","
+ [6] " BASEGEOGCRS[\"Amersfoort\","
+ [7] " DATUM[\"Amersfoort\","
+ [8] " ELLIPSOID[\"Bessel 1841\",6377397.155,299.1528128,"
+ [9] " LENGTHUNIT[\"metre\",1]]],"
+[10] " PRIMEM[\"Greenwich\",0,"
+[11] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+[12] " ID[\"EPSG\",4289]],"
+[13] " CONVERSION[\"RD New\","
+[14] " METHOD[\"Oblique Stereographic\","
+[15] " ID[\"EPSG\",9809]],"
+[16] " PARAMETER[\"Latitude of natural origin\",52.1561605555556,"
+[17] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[18] " ID[\"EPSG\",8801]],"
+[19] " PARAMETER[\"Longitude of natural origin\",5.38763888888889,"
+[20] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[21] " ID[\"EPSG\",8802]],"
+[22] " PARAMETER[\"Scale factor at natural origin\",0.9999079,"
+[23] " SCALEUNIT[\"unity\",1],"
+[24] " ID[\"EPSG\",8805]],"
+[25] " PARAMETER[\"False easting\",155000,"
+[26] " LENGTHUNIT[\"metre\",1],"
+[27] " ID[\"EPSG\",8806]],"
+[28] " PARAMETER[\"False northing\",463000,"
+[29] " LENGTHUNIT[\"metre\",1],"
+[30] " ID[\"EPSG\",8807]]],"
+[31] " CS[Cartesian,2],"
+[32] " AXIS[\"easting (X)\",east,"
+[33] " ORDER[1],"
+[34] " LENGTHUNIT[\"metre\",1]],"
+[35] " AXIS[\"northing (Y)\",north,"
+[36] " ORDER[2],"
+[37] " LENGTHUNIT[\"metre\",1]],"
+[38] " USAGE["
+[39] " SCOPE[\"Engineering survey, topographic mapping.\"],"
+[40] " AREA[\"Netherlands - onshore, including Waddenzee, Dutch Wadden Islands and 12-mile offshore coastal zone.\"],"
+[41] " BBOX[50.75,3.2,53.7,7.22]],"
+[42] " ID[\"EPSG\",28992]]"
+[43] "Data axis to CRS axis mapping: 1,2"
+[44] "Origin = (83565.000000000000000,447180.000000000000000)"
+[45] "Pixel Size = (5.000000000000000,-5.000000000000000)"
+[46] "Metadata:"
+[47] " AREA_OR_POINT=Area"
+[48] "Image Structure Metadata:"
+[49] " INTERLEAVE=BAND"
+[50] "Corner Coordinates:"
+[51] "Upper Left ( 83565.000, 447180.000) ( 4d20'49.32\"E, 52d 0'33.67\"N)"
+[52] "Lower Left ( 83565.000, 445250.000) ( 4d20'50.77\"E, 51d59'31.22\"N)"
+[53] "Upper Right ( 87175.000, 447180.000) ( 4d23'58.60\"E, 52d 0'35.30\"N)"
+[54] "Lower Right ( 87175.000, 445250.000) ( 4d23'59.98\"E, 51d59'32.85\"N)"
+[55] "Center ( 85370.000, 446215.000) ( 4d22'24.67\"E, 52d 0' 3.27\"N)"
+[56] "Band 1 Block=722x2 Type=Float32, ColorInterp=Gray"
+```
+
+We’ve already loaded and worked with these two data files in earlier episodes. Let’s plot them each once more to remind ourselves what this data looks like. First we’ll plot the DTM elevation data:
+
+```r
+ ggplot() +
+ geom_raster(data = DTM_TUD_df ,
+ aes(x = x, y = y, fill = `tud-dtm-5m`)) +
+ scale_fill_gradientn(name = "Elevation", colors = terrain.colors(10)) +
+ coord_quickmap()
+```
+
+
+
+We have now successfully draped the Digital Terrain Model on top of our hillshade to produce a nice looking, textured map!
+
+
+
+::::::::::::::::::::::::::::::::::::: keypoints
+
+- In order to plot two raster data sets together, they must be in the same CRS.
+- Use the `project()` function to convert between CRSs.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
diff --git a/16-raster-calculations.md b/16-raster-calculations.md
new file mode 100644
index 00000000..b2ed9b34
--- /dev/null
+++ b/16-raster-calculations.md
@@ -0,0 +1,324 @@
+---
+title: 'Raster Calculations'
+teaching: 25
+exercises: 5
+---
+
+
+
+:::::::::::::::::::::::::::::::::::::: questions
+
+- How do I subtract one raster from another and extract pixel values for defined locations?
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::::::::::::::::::::::::::::::::::::: objectives
+
+After completing this episode, participants should be able to…
+
+- Perform a subtraction between two rasters using raster math.
+- Export raster data as a GeoTIFF file.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::: prereq
+
+# Things you'll need to complete this episode
+
+See the [setup instructions](../learners/setup.md) for detailed information about the software, data, and other prerequisites you will need to work through the examples in this episode.
+
+
+
+:::
+
+We often want to combine values of and perform calculations on rasters to create a new output raster. This episode covers how to subtract one raster from another using basic raster math.
+
+## Raster calculations in R
+
+We often want to perform calculations on two or more rasters to create a new output raster. For example, if we are interested in mapping the heights of trees and buildings across an entire field site, we might want to calculate the difference between the Digital Surface Model (DSM, tops of trees and buildings) and the Digital Terrain Model (DTM, ground level). The resulting dataset is referred to as a Canopy Height Model (CHM) and represents the actual height of trees, buildings, etc. with the influence of ground elevation removed.
+
+
+
+## Load the Data
+
+For this episode, we will use the DTM and DSM data which we already have loaded from previous episodes.
+
+We use the `describe()` function to view information about the DTM and DSM data files.
+
+```r
+describe("data/tud-dtm-5m.tif")
+```
+
+```{.output}
+ [1] "Driver: GTiff/GeoTIFF"
+ [2] "Files: data/tud-dtm-5m.tif"
+ [3] "Size is 722, 386"
+ [4] "Coordinate System is:"
+ [5] "PROJCRS[\"Amersfoort / RD New\","
+ [6] " BASEGEOGCRS[\"Amersfoort\","
+ [7] " DATUM[\"Amersfoort\","
+ [8] " ELLIPSOID[\"Bessel 1841\",6377397.155,299.1528128,"
+ [9] " LENGTHUNIT[\"metre\",1]]],"
+[10] " PRIMEM[\"Greenwich\",0,"
+[11] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+[12] " ID[\"EPSG\",4289]],"
+[13] " CONVERSION[\"RD New\","
+[14] " METHOD[\"Oblique Stereographic\","
+[15] " ID[\"EPSG\",9809]],"
+[16] " PARAMETER[\"Latitude of natural origin\",52.1561605555556,"
+[17] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[18] " ID[\"EPSG\",8801]],"
+[19] " PARAMETER[\"Longitude of natural origin\",5.38763888888889,"
+[20] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[21] " ID[\"EPSG\",8802]],"
+[22] " PARAMETER[\"Scale factor at natural origin\",0.9999079,"
+[23] " SCALEUNIT[\"unity\",1],"
+[24] " ID[\"EPSG\",8805]],"
+[25] " PARAMETER[\"False easting\",155000,"
+[26] " LENGTHUNIT[\"metre\",1],"
+[27] " ID[\"EPSG\",8806]],"
+[28] " PARAMETER[\"False northing\",463000,"
+[29] " LENGTHUNIT[\"metre\",1],"
+[30] " ID[\"EPSG\",8807]]],"
+[31] " CS[Cartesian,2],"
+[32] " AXIS[\"easting (X)\",east,"
+[33] " ORDER[1],"
+[34] " LENGTHUNIT[\"metre\",1]],"
+[35] " AXIS[\"northing (Y)\",north,"
+[36] " ORDER[2],"
+[37] " LENGTHUNIT[\"metre\",1]],"
+[38] " USAGE["
+[39] " SCOPE[\"Engineering survey, topographic mapping.\"],"
+[40] " AREA[\"Netherlands - onshore, including Waddenzee, Dutch Wadden Islands and 12-mile offshore coastal zone.\"],"
+[41] " BBOX[50.75,3.2,53.7,7.22]],"
+[42] " ID[\"EPSG\",28992]]"
+[43] "Data axis to CRS axis mapping: 1,2"
+[44] "Origin = (83565.000000000000000,447180.000000000000000)"
+[45] "Pixel Size = (5.000000000000000,-5.000000000000000)"
+[46] "Metadata:"
+[47] " AREA_OR_POINT=Area"
+[48] "Image Structure Metadata:"
+[49] " INTERLEAVE=BAND"
+[50] "Corner Coordinates:"
+[51] "Upper Left ( 83565.000, 447180.000) ( 4d20'49.32\"E, 52d 0'33.67\"N)"
+[52] "Lower Left ( 83565.000, 445250.000) ( 4d20'50.77\"E, 51d59'31.22\"N)"
+[53] "Upper Right ( 87175.000, 447180.000) ( 4d23'58.60\"E, 52d 0'35.30\"N)"
+[54] "Lower Right ( 87175.000, 445250.000) ( 4d23'59.98\"E, 51d59'32.85\"N)"
+[55] "Center ( 85370.000, 446215.000) ( 4d22'24.67\"E, 52d 0' 3.27\"N)"
+[56] "Band 1 Block=722x2 Type=Float32, ColorInterp=Gray"
+```
+
+```r
+describe("data/tud-dsm-5m.tif")
+```
+
+```{.output}
+ [1] "Driver: GTiff/GeoTIFF"
+ [2] "Files: data/tud-dsm-5m.tif"
+ [3] "Size is 722, 386"
+ [4] "Coordinate System is:"
+ [5] "PROJCRS[\"Amersfoort / RD New\","
+ [6] " BASEGEOGCRS[\"Amersfoort\","
+ [7] " DATUM[\"Amersfoort\","
+ [8] " ELLIPSOID[\"Bessel 1841\",6377397.155,299.1528128,"
+ [9] " LENGTHUNIT[\"metre\",1]]],"
+[10] " PRIMEM[\"Greenwich\",0,"
+[11] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
+[12] " ID[\"EPSG\",4289]],"
+[13] " CONVERSION[\"RD New\","
+[14] " METHOD[\"Oblique Stereographic\","
+[15] " ID[\"EPSG\",9809]],"
+[16] " PARAMETER[\"Latitude of natural origin\",52.1561605555556,"
+[17] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[18] " ID[\"EPSG\",8801]],"
+[19] " PARAMETER[\"Longitude of natural origin\",5.38763888888889,"
+[20] " ANGLEUNIT[\"degree\",0.0174532925199433],"
+[21] " ID[\"EPSG\",8802]],"
+[22] " PARAMETER[\"Scale factor at natural origin\",0.9999079,"
+[23] " SCALEUNIT[\"unity\",1],"
+[24] " ID[\"EPSG\",8805]],"
+[25] " PARAMETER[\"False easting\",155000,"
+[26] " LENGTHUNIT[\"metre\",1],"
+[27] " ID[\"EPSG\",8806]],"
+[28] " PARAMETER[\"False northing\",463000,"
+[29] " LENGTHUNIT[\"metre\",1],"
+[30] " ID[\"EPSG\",8807]]],"
+[31] " CS[Cartesian,2],"
+[32] " AXIS[\"easting (X)\",east,"
+[33] " ORDER[1],"
+[34] " LENGTHUNIT[\"metre\",1]],"
+[35] " AXIS[\"northing (Y)\",north,"
+[36] " ORDER[2],"
+[37] " LENGTHUNIT[\"metre\",1]],"
+[38] " USAGE["
+[39] " SCOPE[\"Engineering survey, topographic mapping.\"],"
+[40] " AREA[\"Netherlands - onshore, including Waddenzee, Dutch Wadden Islands and 12-mile offshore coastal zone.\"],"
+[41] " BBOX[50.75,3.2,53.7,7.22]],"
+[42] " ID[\"EPSG\",28992]]"
+[43] "Data axis to CRS axis mapping: 1,2"
+[44] "Origin = (83565.000000000000000,447180.000000000000000)"
+[45] "Pixel Size = (5.000000000000000,-5.000000000000000)"
+[46] "Metadata:"
+[47] " AREA_OR_POINT=Area"
+[48] "Image Structure Metadata:"
+[49] " INTERLEAVE=BAND"
+[50] "Corner Coordinates:"
+[51] "Upper Left ( 83565.000, 447180.000) ( 4d20'49.32\"E, 52d 0'33.67\"N)"
+[52] "Lower Left ( 83565.000, 445250.000) ( 4d20'50.77\"E, 51d59'31.22\"N)"
+[53] "Upper Right ( 87175.000, 447180.000) ( 4d23'58.60\"E, 52d 0'35.30\"N)"
+[54] "Lower Right ( 87175.000, 445250.000) ( 4d23'59.98\"E, 51d59'32.85\"N)"
+[55] "Center ( 85370.000, 446215.000) ( 4d22'24.67\"E, 52d 0' 3.27\"N)"
+[56] "Band 1 Block=722x2 Type=Float32, ColorInterp=Gray"
+```
+
+We’ve already loaded and worked with these two data files in earlier episodes. Let’s plot them each once more to remind ourselves what this data looks like. First we’ll plot the DTM elevation data:
+
+```r
+ ggplot() +
+ geom_raster(data = DTM_TUD_df ,
+ aes(x = x, y = y, fill = `tud-dtm-5m`)) +
+ scale_fill_gradientn(name = "Elevation", colors = terrain.colors(10)) +
+ coord_quickmap()
+```
+
+ +
+And then the DSM elevation data:
+
+```r
+ ggplot() +
+ geom_raster(data = DSM_TUD_df ,
+ aes(x = x, y = y, fill = `tud-dsm-5m`)) +
+ scale_fill_gradientn(name = "Elevation", colors = terrain.colors(10)) +
+ coord_quickmap()
+```
+
+
+
+And then the DSM elevation data:
+
+```r
+ ggplot() +
+ geom_raster(data = DSM_TUD_df ,
+ aes(x = x, y = y, fill = `tud-dsm-5m`)) +
+ scale_fill_gradientn(name = "Elevation", colors = terrain.colors(10)) +
+ coord_quickmap()
+```
+
+ +
+## Raster math and Canopy Height Models
+
+We can perform raster calculations by subtracting (or adding, multiplying, etc.) two rasters. In the geospatial world, we call this “raster math”.
+
+Let’s subtract the DTM from the DSM to create a Canopy Height Model. After subtracting, let’s create a data frame so we can plot with `ggplot`.
+
+
+
+```r
+CHM_TUD <- DSM_TUD - DTM_TUD
+CHM_TUD_df <- as.data.frame(CHM_TUD, xy = TRUE)
+```
+
+We can now plot the output CHM.
+
+```r
+ ggplot() +
+ geom_raster(data = CHM_TUD_df ,
+ aes(x = x, y = y, fill = `tud-dsm-5m`)) +
+ scale_fill_gradientn(name = "Canopy Height", colors = terrain.colors(10)) +
+ coord_quickmap()
+```
+
+
+
+## Raster math and Canopy Height Models
+
+We can perform raster calculations by subtracting (or adding, multiplying, etc.) two rasters. In the geospatial world, we call this “raster math”.
+
+Let’s subtract the DTM from the DSM to create a Canopy Height Model. After subtracting, let’s create a data frame so we can plot with `ggplot`.
+
+
+
+```r
+CHM_TUD <- DSM_TUD - DTM_TUD
+CHM_TUD_df <- as.data.frame(CHM_TUD, xy = TRUE)
+```
+
+We can now plot the output CHM.
+
+```r
+ ggplot() +
+ geom_raster(data = CHM_TUD_df ,
+ aes(x = x, y = y, fill = `tud-dsm-5m`)) +
+ scale_fill_gradientn(name = "Canopy Height", colors = terrain.colors(10)) +
+ coord_quickmap()
+```
+
+ +
+Let’s have a look at the distribution of values in our newly created Canopy Height Model (CHM).
+
+```r
+ggplot(CHM_TUD_df) +
+ geom_histogram(aes(`tud-dsm-5m`))
+```
+
+
+
+Let’s have a look at the distribution of values in our newly created Canopy Height Model (CHM).
+
+```r
+ggplot(CHM_TUD_df) +
+ geom_histogram(aes(`tud-dsm-5m`))
+```
+
+ +Notice that the range of values for the output CHM starts right below 0 and ranges to almost 100 meters. Does this make sense for buildings and trees in Delft?
+
+::: challenge
+
+### Challenge: Explore CHM Raster Values
+
+It’s often a good idea to explore the range of values in a raster dataset just like we might explore a dataset that we collected in the field.
+
+1. What is the minimum and maximum value for the Canopy Height Model `CHM_TUD` that we just created?
+2. What is the distribution of all the pixel values in the CHM?
+3. Plot the `CHM_TUD` raster using breaks that make sense for the data. Include an appropriate colour palette for the data, plot title and no axes ticks / labels.
+
+::: solution
+
+
+```r
+min(CHM_TUD_df$`tud-dsm-5m`, na.rm = TRUE)
+```
+
+```{.output}
+[1] -3.638057
+```
+
+```r
+max(CHM_TUD_df$`tud-dsm-5m`, na.rm = TRUE)
+```
+
+```{.output}
+[1] 92.08102
+```
+
+```r
+ggplot(CHM_TUD_df) +
+ geom_histogram(aes(`tud-dsm-5m`))
+```
+
+
+Notice that the range of values for the output CHM starts right below 0 and ranges to almost 100 meters. Does this make sense for buildings and trees in Delft?
+
+::: challenge
+
+### Challenge: Explore CHM Raster Values
+
+It’s often a good idea to explore the range of values in a raster dataset just like we might explore a dataset that we collected in the field.
+
+1. What is the minimum and maximum value for the Canopy Height Model `CHM_TUD` that we just created?
+2. What is the distribution of all the pixel values in the CHM?
+3. Plot the `CHM_TUD` raster using breaks that make sense for the data. Include an appropriate colour palette for the data, plot title and no axes ticks / labels.
+
+::: solution
+
+
+```r
+min(CHM_TUD_df$`tud-dsm-5m`, na.rm = TRUE)
+```
+
+```{.output}
+[1] -3.638057
+```
+
+```r
+max(CHM_TUD_df$`tud-dsm-5m`, na.rm = TRUE)
+```
+
+```{.output}
+[1] 92.08102
+```
+
+```r
+ggplot(CHM_TUD_df) +
+ geom_histogram(aes(`tud-dsm-5m`))
+```
+
+ +
+```r
+custom_bins <- c(-5, 0, 10, 20, 30, 100)
+CHM_TUD_df <- CHM_TUD_df %>%
+ mutate(canopy_discrete = cut(`tud-dsm-5m`, breaks = custom_bins))
+
+ggplot() +
+ geom_raster(data = CHM_TUD_df , aes(x = x, y = y,
+ fill = canopy_discrete)) +
+ scale_fill_manual(values = terrain.colors(5)) +
+ coord_quickmap()
+```
+
+
+
+```r
+custom_bins <- c(-5, 0, 10, 20, 30, 100)
+CHM_TUD_df <- CHM_TUD_df %>%
+ mutate(canopy_discrete = cut(`tud-dsm-5m`, breaks = custom_bins))
+
+ggplot() +
+ geom_raster(data = CHM_TUD_df , aes(x = x, y = y,
+ fill = canopy_discrete)) +
+ scale_fill_manual(values = terrain.colors(5)) +
+ coord_quickmap()
+```
+
+ +
+:::
+
+:::
+
+::: callout
+
+### Two Ways to Perform Raster Calculations
+
+We can calculate the difference between two rasters in two different ways:
+
+- by directly subtracting the two rasters in R using raster math,
+
+or for more efficient processing, particularly if our rasters are large and/or the calculations we are performing are complex:
+
+- using the `lapp()` function.
+
+See how `lapp()` is used in [this lesson](https://datacarpentry.org/r-raster-vector-geospatial/instructor/04-raster-calculations-in-r.html#efficient-raster-calculations).
+
+:::
+
+## Export a GeoTIFF
+
+Now that we’ve created a new raster, let’s export the data as a GeoTIFF file using the `writeRaster()` function.
+
+When we write this raster object to a GeoTIFF file we’ll name it `CHM_TUD.tiff`. This name allows us to quickly remember both what the data contains (CHM data) and for where (TU Delft campus and surroundings). The `writeRaster()` function by default writes the output file to your working directory unless you specify a full file path.
+
+We will specify the output format (“GTiff”), the no data value `NAflag = -9999`. We will also tell R to overwrite any data that is already in a file of the same name.
+
+```r
+writeRaster(CHM_TUD, "fig/CHM_TUD.tiff",
+ filetype="GTiff",
+ overwrite=TRUE,
+ NAflag=-9999)
+```
+
+::::::::::::::::::::::::::::::::::::: keypoints
+
+- Rasters can be computed on using mathematical functions.
+- The `writeRaster()` function can be used to write raster data to a file.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
diff --git a/17-work-with-multi-band-rasters.md b/17-work-with-multi-band-rasters.md
new file mode 100644
index 00000000..e06ec549
--- /dev/null
+++ b/17-work-with-multi-band-rasters.md
@@ -0,0 +1,211 @@
+---
+title: 'Work with Multi-Band Rasters'
+teaching: 25
+exercises: 0
+---
+
+
+
+:::::::::::::::::::::::::::::::::::::: questions
+
+- How can I visualize individual and multiple bands in a raster object?
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::::::::::::::::::::::::::::::::::::: objectives
+
+After completing this episode, participants should be able to…
+
+- Identify a single versus a multi-band raster file.
+- Import multi-band rasters into R using the `terra` package.
+- Plot multi-band colour image rasters in R using the `ggplot` package.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::: prereq
+
+# Things you'll need to complete this episode
+
+See the [setup instructions](../learners/setup.md) for detailed information about the software, data, and other prerequisites you will need to work through the examples in this episode.
+
+
+
+:::
+
+
+This episode explores how to import and plot a multi-band raster in R.
+
+## Getting Started with Multi-Band Data in R
+
+In this episode, the multi-band data that we are working with is [Beeldmateriaal Open Data](https://opendata.beeldmateriaal.nl/) from the Netherlands. Each RGB image is a 3-band raster. The same steps would apply to working with a multi-spectral image with 4 or more bands - like Landsat imagery.
+
+By using the `rast()` function along with the `lyrs` parameter, we can read specific raster bands; omitting this parameter would read instead all bands.
+
+
+```r
+RGB_band1_TUD <- rast("data/tudlib-rgb.tif", lyrs = 1)
+```
+
+We need to convert this data to a data frame in order to plot it with `ggplot`.
+
+```r
+RGB_band1_TUD_df <- as.data.frame(RGB_band1_TUD, xy = TRUE)
+
+ggplot() +
+ geom_raster(data = RGB_band1_TUD_df,
+ aes(x = x, y = y, alpha = `tudlib-rgb_1`)) +
+ coord_equal()
+```
+
+
+
+:::
+
+:::
+
+::: callout
+
+### Two Ways to Perform Raster Calculations
+
+We can calculate the difference between two rasters in two different ways:
+
+- by directly subtracting the two rasters in R using raster math,
+
+or for more efficient processing, particularly if our rasters are large and/or the calculations we are performing are complex:
+
+- using the `lapp()` function.
+
+See how `lapp()` is used in [this lesson](https://datacarpentry.org/r-raster-vector-geospatial/instructor/04-raster-calculations-in-r.html#efficient-raster-calculations).
+
+:::
+
+## Export a GeoTIFF
+
+Now that we’ve created a new raster, let’s export the data as a GeoTIFF file using the `writeRaster()` function.
+
+When we write this raster object to a GeoTIFF file we’ll name it `CHM_TUD.tiff`. This name allows us to quickly remember both what the data contains (CHM data) and for where (TU Delft campus and surroundings). The `writeRaster()` function by default writes the output file to your working directory unless you specify a full file path.
+
+We will specify the output format (“GTiff”), the no data value `NAflag = -9999`. We will also tell R to overwrite any data that is already in a file of the same name.
+
+```r
+writeRaster(CHM_TUD, "fig/CHM_TUD.tiff",
+ filetype="GTiff",
+ overwrite=TRUE,
+ NAflag=-9999)
+```
+
+::::::::::::::::::::::::::::::::::::: keypoints
+
+- Rasters can be computed on using mathematical functions.
+- The `writeRaster()` function can be used to write raster data to a file.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
diff --git a/17-work-with-multi-band-rasters.md b/17-work-with-multi-band-rasters.md
new file mode 100644
index 00000000..e06ec549
--- /dev/null
+++ b/17-work-with-multi-band-rasters.md
@@ -0,0 +1,211 @@
+---
+title: 'Work with Multi-Band Rasters'
+teaching: 25
+exercises: 0
+---
+
+
+
+:::::::::::::::::::::::::::::::::::::: questions
+
+- How can I visualize individual and multiple bands in a raster object?
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::::::::::::::::::::::::::::::::::::: objectives
+
+After completing this episode, participants should be able to…
+
+- Identify a single versus a multi-band raster file.
+- Import multi-band rasters into R using the `terra` package.
+- Plot multi-band colour image rasters in R using the `ggplot` package.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
+::: prereq
+
+# Things you'll need to complete this episode
+
+See the [setup instructions](../learners/setup.md) for detailed information about the software, data, and other prerequisites you will need to work through the examples in this episode.
+
+
+
+:::
+
+
+This episode explores how to import and plot a multi-band raster in R.
+
+## Getting Started with Multi-Band Data in R
+
+In this episode, the multi-band data that we are working with is [Beeldmateriaal Open Data](https://opendata.beeldmateriaal.nl/) from the Netherlands. Each RGB image is a 3-band raster. The same steps would apply to working with a multi-spectral image with 4 or more bands - like Landsat imagery.
+
+By using the `rast()` function along with the `lyrs` parameter, we can read specific raster bands; omitting this parameter would read instead all bands.
+
+
+```r
+RGB_band1_TUD <- rast("data/tudlib-rgb.tif", lyrs = 1)
+```
+
+We need to convert this data to a data frame in order to plot it with `ggplot`.
+
+```r
+RGB_band1_TUD_df <- as.data.frame(RGB_band1_TUD, xy = TRUE)
+
+ggplot() +
+ geom_raster(data = RGB_band1_TUD_df,
+ aes(x = x, y = y, alpha = `tudlib-rgb_1`)) +
+ coord_equal()
+```
+
+ +
+## Image Raster Data Values
+
+This raster contains values between 0 and 255. These values represent degrees of brightness associated with the image band. In the case of a RGB image (red, green and blue), band 1 is the red band. When we plot the red band, larger numbers (towards 255) represent pixels with more red in them (a strong red reflection). Smaller numbers (towards 0) represent pixels with less red in them (less red was reflected). To plot an RGB image, we mix red + green + blue values into one single colour to create a full colour image - similar to the colour image a digital camera creates.
+
+## Import a Specific Band
+
+We can use the `rast()` function to import specific bands in our raster object by specifying which band we want with `lyrs = N` (N represents the band number we want to work with). To import the green band, we would use `lyrs = 2`.
+
+
+```r
+RGB_band2_TUD <- rast("data/tudlib-rgb.tif", lyrs = 2)
+```
+
+We can convert this data to a data frame and plot the same way we plotted the red band:
+
+```r
+RGB_band2_TUD_df <- as.data.frame(RGB_band2_TUD, xy = TRUE)
+
+ggplot() +
+ geom_raster(data = RGB_band2_TUD_df,
+ aes(x = x, y = y, alpha = `tudlib-rgb_2`)) +
+ coord_equal()
+```
+
+
+
+## Image Raster Data Values
+
+This raster contains values between 0 and 255. These values represent degrees of brightness associated with the image band. In the case of a RGB image (red, green and blue), band 1 is the red band. When we plot the red band, larger numbers (towards 255) represent pixels with more red in them (a strong red reflection). Smaller numbers (towards 0) represent pixels with less red in them (less red was reflected). To plot an RGB image, we mix red + green + blue values into one single colour to create a full colour image - similar to the colour image a digital camera creates.
+
+## Import a Specific Band
+
+We can use the `rast()` function to import specific bands in our raster object by specifying which band we want with `lyrs = N` (N represents the band number we want to work with). To import the green band, we would use `lyrs = 2`.
+
+
+```r
+RGB_band2_TUD <- rast("data/tudlib-rgb.tif", lyrs = 2)
+```
+
+We can convert this data to a data frame and plot the same way we plotted the red band:
+
+```r
+RGB_band2_TUD_df <- as.data.frame(RGB_band2_TUD, xy = TRUE)
+
+ggplot() +
+ geom_raster(data = RGB_band2_TUD_df,
+ aes(x = x, y = y, alpha = `tudlib-rgb_2`)) +
+ coord_equal()
+```
+
+ +
+## Raster Stacks
+
+Next, we will work with all three image bands (red, green and blue) as an R raster object. We will then plot a 3-band composite, or full-colour, image.
+
+To bring in all bands of a multi-band raster, we use the `rast()` function.
+
+```r
+RGB_stack_TUD <- rast("data/tudlib-rgb.tif")
+```
+
+Let’s preview the attributes of our stack object:
+
+```r
+RGB_stack_TUD
+```
+
+```{.output}
+class : SpatRaster
+dimensions : 4988, 4866, 3 (nrow, ncol, nlyr)
+resolution : 0.08, 0.08 (x, y)
+extent : 85272, 85661.28, 446295.2, 446694.2 (xmin, xmax, ymin, ymax)
+coord. ref. : Amersfoort / RD New (EPSG:28992)
+source : tudlib-rgb.tif
+colors RGB : 1, 2, 3
+names : tudlib-rgb_1, tudlib-rgb_2, tudlib-rgb_3
+```
+We can view the attributes of each band in the stack in a single output. For example, if we had hundreds of bands, we could specify which band we’d like to view attributes for using an index value:
+
+```r
+RGB_stack_TUD[[2]]
+```
+
+```{.output}
+class : SpatRaster
+dimensions : 4988, 4866, 1 (nrow, ncol, nlyr)
+resolution : 0.08, 0.08 (x, y)
+extent : 85272, 85661.28, 446295.2, 446694.2 (xmin, xmax, ymin, ymax)
+coord. ref. : Amersfoort / RD New (EPSG:28992)
+source : tudlib-rgb.tif
+name : tudlib-rgb_2
+```
+We can also use `ggplot2` to plot the data in any layer of our raster object. Remember, we need to convert to a data frame first.
+
+
+```r
+RGB_stack_TUD_df <- as.data.frame(RGB_stack_TUD, xy = TRUE)
+```
+
+Each band in our RasterStack gets its own column in the data frame. Thus we have:
+
+```r
+str(RGB_stack_TUD_df)
+```
+
+```{.output}
+'data.frame': 24271608 obs. of 5 variables:
+ $ x : num 85272 85272 85272 85272 85272 ...
+ $ y : num 446694 446694 446694 446694 446694 ...
+ $ tudlib-rgb_1: int 52 48 47 49 47 45 47 48 49 54 ...
+ $ tudlib-rgb_2: int 64 58 57 60 57 55 58 59 62 69 ...
+ $ tudlib-rgb_3: int 57 49 49 53 50 47 51 54 57 68 ...
+```
+Let’s create a histogram of the first band:
+
+```r
+ggplot() +
+ geom_histogram(data = RGB_stack_TUD_df, aes(`tudlib-rgb_1`))
+```
+
+
+
+## Raster Stacks
+
+Next, we will work with all three image bands (red, green and blue) as an R raster object. We will then plot a 3-band composite, or full-colour, image.
+
+To bring in all bands of a multi-band raster, we use the `rast()` function.
+
+```r
+RGB_stack_TUD <- rast("data/tudlib-rgb.tif")
+```
+
+Let’s preview the attributes of our stack object:
+
+```r
+RGB_stack_TUD
+```
+
+```{.output}
+class : SpatRaster
+dimensions : 4988, 4866, 3 (nrow, ncol, nlyr)
+resolution : 0.08, 0.08 (x, y)
+extent : 85272, 85661.28, 446295.2, 446694.2 (xmin, xmax, ymin, ymax)
+coord. ref. : Amersfoort / RD New (EPSG:28992)
+source : tudlib-rgb.tif
+colors RGB : 1, 2, 3
+names : tudlib-rgb_1, tudlib-rgb_2, tudlib-rgb_3
+```
+We can view the attributes of each band in the stack in a single output. For example, if we had hundreds of bands, we could specify which band we’d like to view attributes for using an index value:
+
+```r
+RGB_stack_TUD[[2]]
+```
+
+```{.output}
+class : SpatRaster
+dimensions : 4988, 4866, 1 (nrow, ncol, nlyr)
+resolution : 0.08, 0.08 (x, y)
+extent : 85272, 85661.28, 446295.2, 446694.2 (xmin, xmax, ymin, ymax)
+coord. ref. : Amersfoort / RD New (EPSG:28992)
+source : tudlib-rgb.tif
+name : tudlib-rgb_2
+```
+We can also use `ggplot2` to plot the data in any layer of our raster object. Remember, we need to convert to a data frame first.
+
+
+```r
+RGB_stack_TUD_df <- as.data.frame(RGB_stack_TUD, xy = TRUE)
+```
+
+Each band in our RasterStack gets its own column in the data frame. Thus we have:
+
+```r
+str(RGB_stack_TUD_df)
+```
+
+```{.output}
+'data.frame': 24271608 obs. of 5 variables:
+ $ x : num 85272 85272 85272 85272 85272 ...
+ $ y : num 446694 446694 446694 446694 446694 ...
+ $ tudlib-rgb_1: int 52 48 47 49 47 45 47 48 49 54 ...
+ $ tudlib-rgb_2: int 64 58 57 60 57 55 58 59 62 69 ...
+ $ tudlib-rgb_3: int 57 49 49 53 50 47 51 54 57 68 ...
+```
+Let’s create a histogram of the first band:
+
+```r
+ggplot() +
+ geom_histogram(data = RGB_stack_TUD_df, aes(`tudlib-rgb_1`))
+```
+
+ +And a raster plot of the second band:
+
+```r
+ggplot() +
+ geom_raster(data = RGB_stack_TUD_df,
+ aes(x = x, y = y, alpha = `tudlib-rgb_2`)) +
+ coord_equal()
+```
+
+
+And a raster plot of the second band:
+
+```r
+ggplot() +
+ geom_raster(data = RGB_stack_TUD_df,
+ aes(x = x, y = y, alpha = `tudlib-rgb_2`)) +
+ coord_equal()
+```
+
+ +
+We can access any individual band in the same way.
+
+## Create a three-band image
+
+To render a final three-band, coloured image in R, we use the `plotRGB()` function.
+
+This function allows us to:
+
+- Identify what bands we want to render in the red, green and blue regions. The `plotRGB()` function defaults to a 1=red, 2=green, and 3=blue band order. However, you can define what bands you’d like to plot manually. Manual definition of bands is useful if you have, for example a near-infrared band and want to create a colour infrared image.
+- Adjust the stretch of the image to increase or decrease contrast.
+
+Let’s plot our 3-band image. Note that we can use the `plotRGB()` function directly with our RasterStack object (we don’t need a data frame as this function isn’t part of the `ggplot2` package).
+
+
+```r
+plotRGB(RGB_stack_TUD,
+ r = 1, g = 2, b = 3)
+```
+
+
+
+We can access any individual band in the same way.
+
+## Create a three-band image
+
+To render a final three-band, coloured image in R, we use the `plotRGB()` function.
+
+This function allows us to:
+
+- Identify what bands we want to render in the red, green and blue regions. The `plotRGB()` function defaults to a 1=red, 2=green, and 3=blue band order. However, you can define what bands you’d like to plot manually. Manual definition of bands is useful if you have, for example a near-infrared band and want to create a colour infrared image.
+- Adjust the stretch of the image to increase or decrease contrast.
+
+Let’s plot our 3-band image. Note that we can use the `plotRGB()` function directly with our RasterStack object (we don’t need a data frame as this function isn’t part of the `ggplot2` package).
+
+
+```r
+plotRGB(RGB_stack_TUD,
+ r = 1, g = 2, b = 3)
+```
+
+ +
+The image above looks pretty good. We can explore whether applying a stretch to the image might improve clarity and contrast using stretch="lin" or stretch="hist", as explained in [this lesson](https://datacarpentry.org/r-raster-vector-geospatial/instructor/05-raster-multi-band-in-r.html#create-a-three-band-image).
+
+::: callout
+
+### SpatRaster in R
+
+The R SpatRaster object type can handle rasters with multiple bands. The SpatRaster only holds parameters that describe the properties of raster data that is located somewhere on our computer.
+
+A SpatRasterDataset object can hold references to sub-datasets, that is, SpatRaster objects. In most cases, we can work with a SpatRaster in the same way we might work with a SpatRasterDataset.
+
+Read more about SpatRasters in [this lesson](https://datacarpentry.org/r-raster-vector-geospatial/instructor/05-raster-multi-band-in-r.html#spatraster-in-r).
+
+:::
+
+::::::::::::::::::::::::::::::::::::: keypoints
+
+- A single raster file can contain multiple bands or layers.
+- Use the `rast()` function to load all bands in a multi-layer raster file into R.
+- Individual bands within a SpatRaster can be accessed, analysed, and visualized using the same functions no matter how many bands it holds.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
diff --git a/3-raster-slides.html b/3-raster-slides.html
new file mode 100644
index 00000000..543160c6
--- /dev/null
+++ b/3-raster-slides.html
@@ -0,0 +1,836 @@
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+The image above looks pretty good. We can explore whether applying a stretch to the image might improve clarity and contrast using stretch="lin" or stretch="hist", as explained in [this lesson](https://datacarpentry.org/r-raster-vector-geospatial/instructor/05-raster-multi-band-in-r.html#create-a-three-band-image).
+
+::: callout
+
+### SpatRaster in R
+
+The R SpatRaster object type can handle rasters with multiple bands. The SpatRaster only holds parameters that describe the properties of raster data that is located somewhere on our computer.
+
+A SpatRasterDataset object can hold references to sub-datasets, that is, SpatRaster objects. In most cases, we can work with a SpatRaster in the same way we might work with a SpatRasterDataset.
+
+Read more about SpatRasters in [this lesson](https://datacarpentry.org/r-raster-vector-geospatial/instructor/05-raster-multi-band-in-r.html#spatraster-in-r).
+
+:::
+
+::::::::::::::::::::::::::::::::::::: keypoints
+
+- A single raster file can contain multiple bands or layers.
+- Use the `rast()` function to load all bands in a multi-layer raster file into R.
+- Individual bands within a SpatRaster can be accessed, analysed, and visualized using the same functions no matter how many bands it holds.
+
+::::::::::::::::::::::::::::::::::::::::::::::::
+
diff --git a/3-raster-slides.html b/3-raster-slides.html
new file mode 100644
index 00000000..543160c6
--- /dev/null
+++ b/3-raster-slides.html
@@ -0,0 +1,836 @@
+
+
+
+
+
+
+
+
+
+
+
+
+
+