diff --git a/.github/workflows/test_and_deploy.yml b/.github/workflows/test_and_deploy.yml
new file mode 100644
index 00000000..3105dc00
--- /dev/null
+++ b/.github/workflows/test_and_deploy.yml
@@ -0,0 +1,107 @@
+name: tests
+
+on:
+ push:
+ branches:
+ - "main"
+ tags:
+ - "*"
+ pull_request:
+ workflow_dispatch:
+ inputs:
+ coredev:
+ description: 'Tests with development version of cellfinder-core?'
+ required: true
+ default: 'false'
+
+env:
+ REGISTRY: ghcr.io
+ IMAGE_NAME: ${{ github.repository }}
+
+jobs:
+ linting:
+ name: Linting
+ runs-on: ubuntu-latest
+ steps:
+ - uses: neuroinformatics-unit/actions/lint@v2
+
+ manifest:
+ name: Check manifest
+ runs-on: ubuntu-latest
+ steps:
+ - uses: neuroinformatics-unit/actions/check_manifest@v2
+
+ test:
+ needs: [linting,manifest]
+ runs-on: ${{ matrix.os }}
+ env:
+ INPUT_COREDEV: ${{ github.event.inputs.coredev }}
+ strategy:
+ fail-fast: false
+ matrix:
+ # Run across a mixture of Python versions and operating systems
+ include:
+ - os: macos-latest
+ python-version: "3.10"
+ - os: windows-latest
+ python-version: "3.9"
+ - os: ubuntu-latest
+ python-version: "3.8"
+ steps:
+ - uses: neuroinformatics-unit/actions/test@v2
+ with:
+ python-version: ${{ matrix.python-version }}
+
+ build_sdist_wheels:
+ name: Build source distribution
+ needs: [test]
+ if: github.event_name == 'push' && github.ref_type == 'tag'
+ runs-on: ubuntu-latest
+ steps:
+ - uses: neuroinformatics-unit/actions/build_sdist_wheels@v2
+
+ deploy:
+ needs: [build_sdist_wheels]
+ runs-on: ubuntu-latest
+ if: github.event_name == 'push' && github.ref_type == 'tag'
+ steps:
+ - uses: actions/download-artifact@v3
+ with:
+ name: artifact
+ path: dist
+ - uses: pypa/gh-action-pypi-publish@v1.5.0
+ with:
+ user: __token__
+ password: ${{ secrets.TWINE_API_KEY }}
+
+ build-and-push-docker:
+ needs: deploy
+ runs-on: ubuntu-latest
+ permissions:
+ contents: read
+ packages: write
+
+ steps:
+ - name: Checkout repository
+ uses: actions/checkout@v2
+
+ - name: Log in to the Container registry
+ uses: docker/login-action@f054a8b539a109f9f41c372932f1ae047eff08c9
+ with:
+ registry: ${{ env.REGISTRY }}
+ username: ${{ github.actor }}
+ password: ${{ secrets.GITHUB_TOKEN }}
+
+ - name: Extract metadata (tags, labels) for Docker
+ id: meta
+ uses: docker/metadata-action@98669ae865ea3cffbcbaa878cf57c20bbf1c6c38
+ with:
+ images: ${{ env.REGISTRY }}/${{ env.IMAGE_NAME }}
+
+ - name: Build and push Docker image
+ uses: docker/build-push-action@ad44023a93711e3deb337508980b4b5e9bcdc5dc
+ with:
+ context: .
+ push: true
+ tags: ${{ steps.meta.outputs.tags }}
+ labels: ${{ steps.meta.outputs.labels }}
diff --git a/.gitignore b/.gitignore
new file mode 100644
index 00000000..8c1f8334
--- /dev/null
+++ b/.gitignore
@@ -0,0 +1,126 @@
+# Custom config files
+*.conf.custom
+
+# Byte-compiled / optimized / DLL files
+**/__pycache__/**
+*.py[cod]
+*$py.class
+
+# Cython
+*.c
+*.cpp
+
+# C extensions
+*.so
+
+# Distribution / packaging
+.Python
+build/
+develop-eggs/
+dist/
+downloads/
+eggs/
+.eggs/
+lib/
+lib64/
+parts/
+sdist/
+var/
+wheels/
+*.egg-info/
+.installed.cfg
+*.egg
+MANIFEST
+
+# PyInstaller
+# Usually these files are written by a python script from a template
+# before PyInstaller builds the exe, so as to inject date/other infos into it.
+*.manifest
+*.spec
+
+# Installer logs
+pip-log.txt
+pip-delete-this-directory.txt
+
+# Unit test / coverage reports
+htmlcov/
+.tox/
+.coverage
+.coverage.*
+.cache
+nosetests.xml
+coverage.xml
+*.cover
+.hypothesis/
+.pytest_cache/
+
+# Translations
+*.mo
+*.pot
+
+# Django stuff:
+*.log
+local_settings.py
+db.sqlite3
+
+# Flask stuff:
+instance/
+.webassets-cache
+
+# Scrapy stuff:
+.scrapy
+
+# Sphinx documentation
+doc/build/
+
+# pydocmd
+_build/
+mkdocs.yml
+
+# PyBuilder
+target/
+
+# Jupyter Notebook
+.ipynb_checkpoints
+
+# pyenv
+.python-version
+
+# celery beat schedule file
+celerybeat-schedule
+
+# SageMath parsed files
+*.sage.py
+
+# Environments
+.env
+.venv
+env/
+venv/
+ENV/
+env.bak/
+venv.bak/
+.envrc
+
+# Spyder project settings
+.spyderproject
+.spyproject
+
+# Rope project settings
+.ropeproject
+
+# mkdocs documentation
+/site
+
+# mypy
+.mypy_cache/
+
+.idea/
+
+*.~lock.*
+
+pip-wheel-metadata/
+
+*.DS_Store
+
+*.vscode/
diff --git a/.pre-commit-config.yaml b/.pre-commit-config.yaml
new file mode 100644
index 00000000..f60812bd
--- /dev/null
+++ b/.pre-commit-config.yaml
@@ -0,0 +1,32 @@
+# Configuring https://pre-commit.ci/
+ci:
+ autoupdate_schedule: monthly
+
+repos:
+ - repo: https://github.com/pre-commit/pre-commit-hooks
+ rev: v4.4.0
+ hooks:
+ - id: check-docstring-first
+ - id: check-executables-have-shebangs
+ - id: check-merge-conflict
+ - id: check-toml
+ - id: end-of-file-fixer
+ - id: mixed-line-ending
+ args: [--fix=lf]
+ - id: requirements-txt-fixer
+ - id: trailing-whitespace
+ - repo: https://github.com/astral-sh/ruff-pre-commit
+ rev: v0.0.286
+ hooks:
+ - id: ruff
+ - repo: https://github.com/psf/black
+ rev: 23.7.0
+ hooks:
+ - id: black
+ - repo: https://github.com/pre-commit/mirrors-mypy
+ rev: v1.5.1
+ hooks:
+ - id: mypy
+ additional_dependencies:
+ - types-setuptools
+ - types-requests
diff --git a/Dockerfile b/Dockerfile
new file mode 100644
index 00000000..f8393006
--- /dev/null
+++ b/Dockerfile
@@ -0,0 +1,4 @@
+FROM tensorflow/tensorflow:latest-gpu
+LABEL maintainer="code@adamltyson.com"
+RUN pip install cellfinder
+CMD ["bash"]
diff --git a/LICENSE b/LICENSE
new file mode 100644
index 00000000..3e221c2a
--- /dev/null
+++ b/LICENSE
@@ -0,0 +1,29 @@
+BSD 3-Clause License
+
+Copyright (c) 2020, University College London
+All rights reserved.
+
+Redistribution and use in source and binary forms, with or without
+modification, are permitted provided that the following conditions are met:
+
+* Redistributions of source code must retain the above copyright notice, this
+ list of conditions and the following disclaimer.
+
+* Redistributions in binary form must reproduce the above copyright notice,
+ this list of conditions and the following disclaimer in the documentation
+ and/or other materials provided with the distribution.
+
+* Neither the name of the copyright holder nor the names of its
+ contributors may be used to endorse or promote products derived from
+ this software without specific prior written permission.
+
+THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
+AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
+IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE
+DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE
+FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL
+DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR
+SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER
+CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY,
+OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE
+OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
diff --git a/MANIFEST.in b/MANIFEST.in
new file mode 100644
index 00000000..c7354c00
--- /dev/null
+++ b/MANIFEST.in
@@ -0,0 +1,18 @@
+include README.md
+include LICENSE
+
+exclude *.yaml
+exclude *.yml
+exclude Dockerfile
+exclude *.ini
+
+recursive-include cellfinder *.py
+include cellfinder/config/*
+
+global-include *.pxd
+
+prune tests
+prune resources
+
+prune .github
+prune .tox
diff --git a/README.md b/README.md
index e69de29b..b831d0dc 100644
--- a/README.md
+++ b/README.md
@@ -0,0 +1,150 @@
+[](https://pypi.org/project/cellfinder)
+[](https://pypi.org/project/cellfinder)
+[](https://pepy.tech/project/cellfinder)
+[](https://pypi.org/project/cellfinder)
+[](https://github.com/brainglobe/cellfinder)
+[](
+ https://github.com/brainglobe/cellfinder/actions)
+[](https://codecov.io/gh/brainglobe/cellfinder)
+[](https://github.com/python/black)
+[](https://pycqa.github.io/isort/)
+[](https://github.com/pre-commit/pre-commit)
+[](https://docs.brainglobe.info/cellfinder/contributing)
+[](https://brainglobe.info/documentation/cellfinder/index.html)
+[](https://twitter.com/brain_globe)
+
+# Cellfinder
+Whole-brain cell detection, registration and analysis.
+
+**N.B. If you want to just use the cell detection part of cellfinder, please
+see the standalone [cellfinder-core](https://github.com/brainglobe/cellfinder-core)
+package, or the [cellfinder plugin](https://github.com/brainglobe/cellfinder-napari)
+for [napari](https://napari.org/).**
+
+---
+`cellfinder` is a collection of tools developed by [Adam Tyson](https://github.com/adamltyson), [Charly Rousseau](https://github.com/crousseau) and [Christian Niedworok](https://github.com/cniedwor) in the [Margrie Lab](https://www.sainsburywellcome.org/web/groups/margrie-lab), generously supported by the [Sainsbury Wellcome Centre](https://www.sainsburywellcome.org/web/).
+
+`cellfinder` is a designed for the analysis of whole-brain imaging data such as
+ [serial-section imaging](https://sainsburywellcomecentre.github.io/OpenSerialSection/)
+ and lightsheet imaging in cleared tissue. The aim is to provide a single solution for:
+
+ * Cell detection (initial cell candidate detection and refinement using
+ deep learning) (using [cellfinder-core](https://github.com/brainglobe/cellfinder-core))
+ * Atlas registration (using [brainreg](https://github.com/brainglobe/brainreg))
+ * Analysis of cell positions in a common space
+
+ ---
+Installation is with
+`pip install cellfinder`
+
+---
+Basic usage:
+```bash
+cellfinder -s signal_images -b background_images -o output_dir --metadata metadata
+```
+Full documentation can be
+found [here](https://brainglobe.info/documentation/cellfinder/index.html).
+
+This software is at a very early stage, and was written with our data in mind.
+Over time we hope to support other data types/formats. If you have any issues, please get in touch [on the forum](https://forum.image.sc/tag/brainglobe) or by
+[raising an issue](https://github.com/brainglobe/cellfinder/issues/new/choose).
+
+
+---
+## Illustration
+
+### Introduction
+cellfinder takes a stitched, but otherwise raw whole-brain dataset with at least
+two channels:
+ * Background channel (i.e. autofluorescence)
+ * Signal channel, the one with the cells to be detected:
+
+
+**Raw coronal serial two-photon mouse brain image showing labelled cells**
+
+
+### Cell candidate detection
+Classical image analysis (e.g. filters, thresholding) is used to find
+cell-like objects (with false positives):
+
+
+**Candidate cells (including many artefacts)**
+
+
+### Cell candidate classification
+A deep-learning network (ResNet) is used to classify cell candidates as true
+cells or artefacts:
+
+
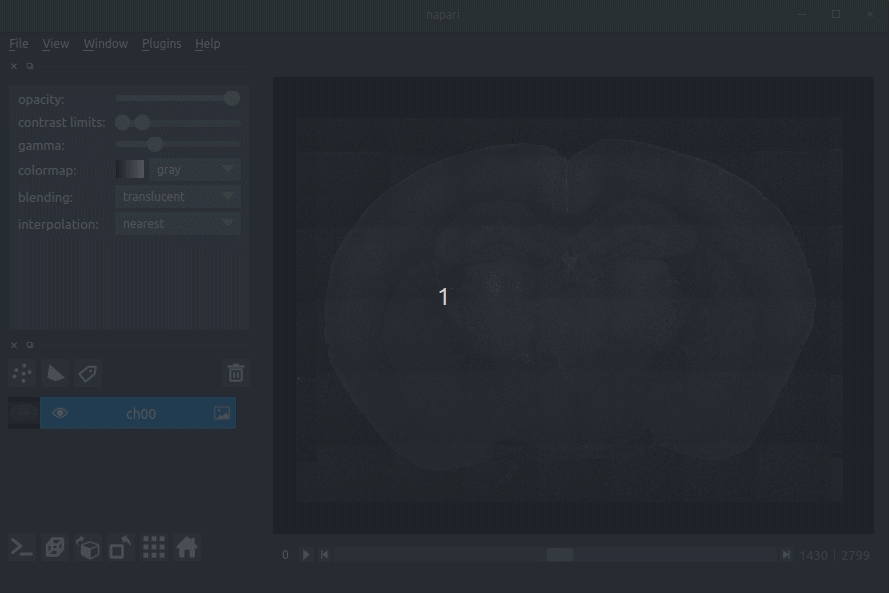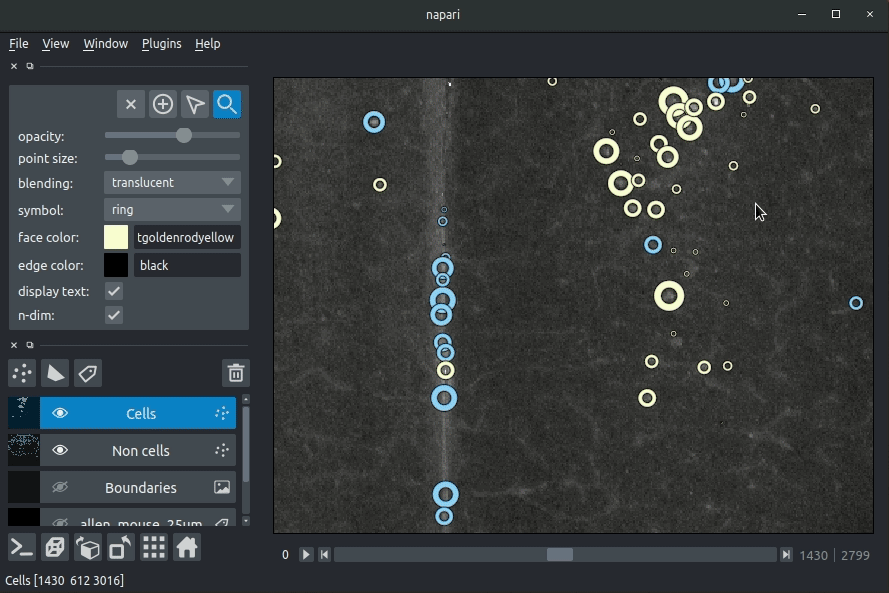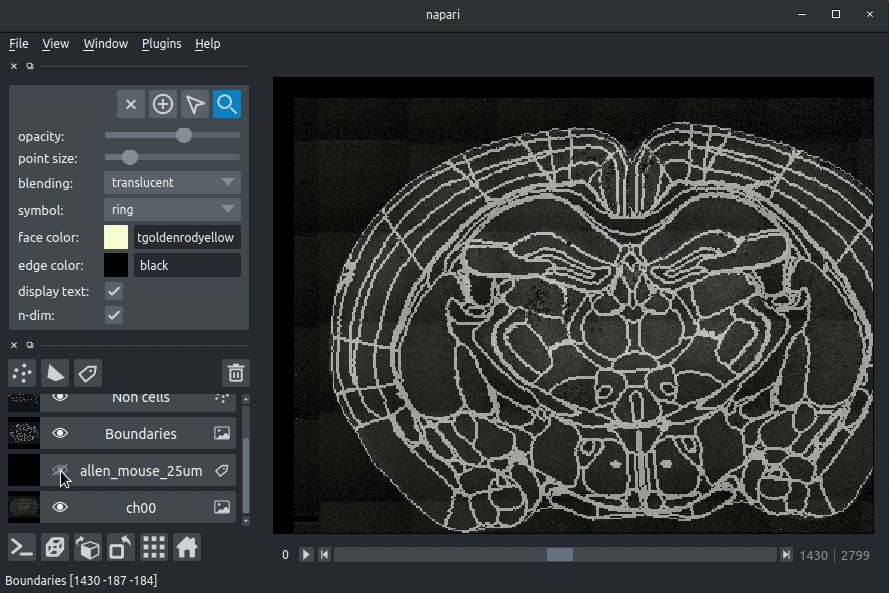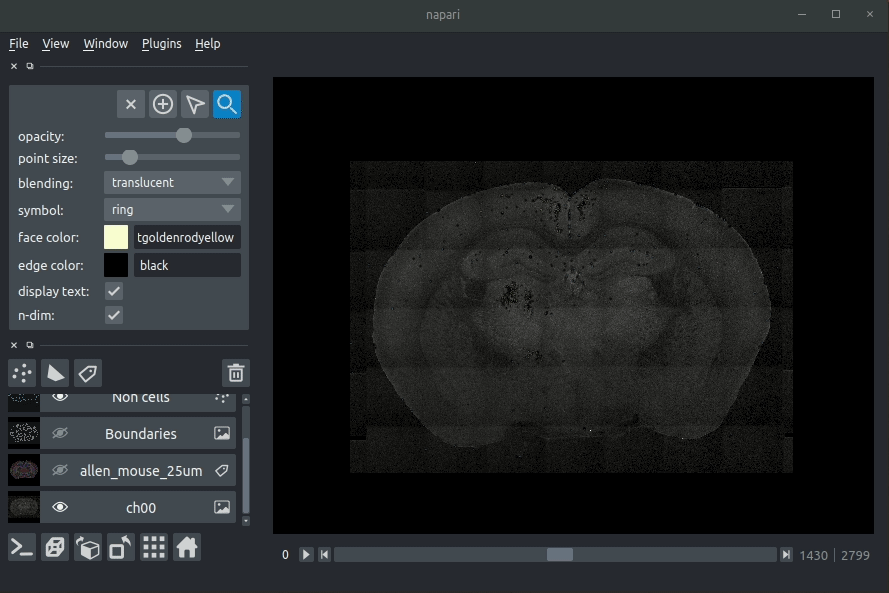
+**Cassified cell candidates. Yellow - cells, Blue - artefacts**
+
+### Registration and segmentation (brainreg)
+Using [brainreg](https://github.com/brainglobe/brainreg),
+cellfinder aligns a template brain and atlas annotations (e.g.
+the Allen Reference Atlas, ARA) to the sample allowing detected cells to be assigned
+a brain region.
+
+This transformation can be inverted, allowing detected cells to be
+transformed to a standard anatomical space.
+
+
+**ARA overlaid on sample image**
+
+### Analysis of cell positions in a common anatomical space
+Registration to a template allows for powerful group-level analysis of cellular
+disributions. *(Example to come)*
+
+## Examples
+*(more to come)*
+
+### Tracing of inputs to retrosplenial cortex (RSP)
+Input cell somas detected by cellfinder, aligned to the Allen Reference Atlas,
+and visualised in [brainrender](https://github.com/brainglobe/brainrender) along
+with RSP.
+
+
+
+Data courtesy of Sepiedeh Keshavarzi and Chryssanthi Tsitoura. [Details here](https://www.youtube.com/watch?v=pMHP0o-KsoQ)
+
+## Visualisation
+
+cellfinder comes with a plugin ([brainglobe-napari-io](https://github.com/brainglobe/brainglobe-napari-io)) for [napari](https://github.com/napari/napari) to view your data
+
+#### Usage
+* Open napari (however you normally do it, but typically just type `napari` into your terminal, or click on your desktop icon)
+
+#### Load cellfinder XML file
+* Load your raw data (drag and drop the data directories into napari, one at a time)
+* Drag and drop your cellfinder XML file (e.g. `cell_classification.xml`) into napari.
+
+#### Load cellfinder directory
+* Load your raw data (drag and drop the data directories into napari, one at a time)
+* Drag and drop your cellfinder output directory into napari.
+
+The plugin will then load your detected cells (in yellow) and the rejected cell
+candidates (in blue). If you carried out registration, then these results will be
+overlaid (similarly to the loading brainreg data, but transformed to the
+coordinate space of your raw data).
+
+
+**Loading raw data**
+
+
+**Loading cellfinder results**
+
+
+## Contributing
+Contributions to cellfinder are more than welcome. Please see the [developers guide](https://brainglobe.info/developers/index.html).
+
+
+## Citing cellfinder
+
+If you find cellfinder useful, and use it in your research, please cite the paper outlining the cell detection algorithm:
+> Tyson, A. L., Rousseau, C. V., Niedworok, C. J., Keshavarzi, S., Tsitoura, C., Cossell, L., Strom, M. and Margrie, T. W. (2021) “A deep learning algorithm for 3D cell detection in whole mouse brain image datasets’ PLOS Computational Biology, 17(5), e1009074
+[https://doi.org/10.1371/journal.pcbi.1009074](https://doi.org/10.1371/journal.pcbi.1009074)
+>
+If you use any of the image registration functions in cellfinder, please also cite [brainreg](https://github.com/brainglobe/brainreg#citing-brainreg).
+
+**If you use this, or any other tools in the brainglobe suite, please
+ [let us know](mailto:code@adamltyson.com?subject=cellfinder), and
+ we'd be happy to promote your paper/talk etc.**
diff --git a/cellfinder/__init__.py b/cellfinder/__init__.py
new file mode 100644
index 00000000..dece5176
--- /dev/null
+++ b/cellfinder/__init__.py
@@ -0,0 +1,7 @@
+from importlib.metadata import metadata
+
+__version__ = metadata("cellfinder")["version"]
+__author__ = metadata("cellfinder")["author-email"]
+__license__ = metadata("cellfinder")["license"]
+
+del metadata
diff --git a/cellfinder/analyse/__init__.py b/cellfinder/analyse/__init__.py
new file mode 100644
index 00000000..e69de29b
diff --git a/cellfinder/analyse/analyse.py b/cellfinder/analyse/analyse.py
new file mode 100644
index 00000000..17133241
--- /dev/null
+++ b/cellfinder/analyse/analyse.py
@@ -0,0 +1,368 @@
+"""
+Cell position analysis (based on atlas registration).
+
+Based on https://github.com/SainsburyWellcomeCentre/cell_count_analysis by
+Charly Rousseau (https://github.com/crousseau).
+"""
+import logging
+import os
+from pathlib import Path
+from typing import List, Optional
+
+import bg_space as bgs
+import imio
+import numpy as np
+import pandas as pd
+import tifffile
+from bg_atlasapi import BrainGlobeAtlas
+from brainglobe_utils.general.system import ensure_directory_exists
+from brainglobe_utils.pandas.misc import sanitise_df
+
+from cellfinder.export.export import export_points
+
+
+class Point:
+ def __init__(
+ self,
+ raw_coordinate,
+ atlas_coordinate,
+ structure,
+ structure_id,
+ hemisphere,
+ ):
+ self.raw_coordinate = raw_coordinate
+ self.atlas_coordinate = atlas_coordinate
+ self.structure = structure
+ self.structure_id = structure_id
+ self.hemisphere = hemisphere
+
+
+def calculate_densities(counts, volume_csv_path):
+ """
+ Use the region volume information from registration to calculate cell
+ densities. Based on the atlas names, which must be exactly equal.
+ :param counts: dataframe with cell counts
+ :param volume_csv_path: path of the volumes of each brain region
+ :return:
+ """
+ volumes = pd.read_csv(volume_csv_path, sep=",", header=0, quotechar='"')
+ df = pd.merge(counts, volumes, on="structure_name", how="outer")
+ df = df.fillna(0)
+ df["left_cells_per_mm3"] = df.left_cell_count / df.left_volume_mm3
+ df["right_cells_per_mm3"] = df.right_cell_count / df.right_volume_mm3
+ return df
+
+
+def combine_df_hemispheres(df):
+ """
+ Combine left and right hemisphere data onto a single row
+ :param df:
+ :return:
+ """
+ left = df[df["hemisphere"] == "left"]
+ right = df[df["hemisphere"] == "right"]
+ left = left.drop(["hemisphere"], axis=1)
+ right = right.drop(["hemisphere"], axis=1)
+ left.rename(columns={"cell_count": "left_cell_count"}, inplace=True)
+ right.rename(columns={"cell_count": "right_cell_count"}, inplace=True)
+ both = pd.merge(left, right, on="structure_name", how="outer")
+ both = both.fillna(0)
+ both["total_cells"] = both.left_cell_count + both.right_cell_count
+ both = both.sort_values("total_cells", ascending=False)
+ return both
+
+
+def summarise_points(
+ raw_points,
+ transformed_points,
+ atlas,
+ volume_csv_path,
+ all_points_filename,
+ summary_filename,
+):
+ points = []
+ structures_with_points = set()
+ for idx, point in enumerate(transformed_points):
+ try:
+ structure_id = atlas.structure_from_coords(point)
+ structure = atlas.structures[structure_id]["name"]
+ hemisphere = atlas.hemisphere_from_coords(point, as_string=True)
+ points.append(
+ Point(
+ raw_points[idx], point, structure, structure_id, hemisphere
+ )
+ )
+ structures_with_points.add(structure)
+ except Exception:
+ continue
+
+ logging.debug("Ensuring output directory exists")
+ ensure_directory_exists(Path(all_points_filename).parent)
+ create_all_cell_csv(points, all_points_filename)
+
+ get_region_totals(
+ points, structures_with_points, volume_csv_path, summary_filename
+ )
+
+ return points
+
+
+def create_all_cell_csv(points, all_points_filename):
+ df = pd.DataFrame(
+ columns=(
+ "coordinate_raw_axis_0",
+ "coordinate_raw_axis_1",
+ "coordinate_raw_axis_2",
+ "coordinate_atlas_axis_0",
+ "coordinate_atlas_axis_1",
+ "coordinate_atlas_axis_2",
+ "structure_name",
+ "hemisphere",
+ )
+ )
+
+ temp_matrix = [[] for i in range(len(points))]
+ for i, point in enumerate(points):
+ temp_matrix[i].append(point.raw_coordinate[0])
+ temp_matrix[i].append(point.raw_coordinate[1])
+ temp_matrix[i].append(point.raw_coordinate[2])
+ temp_matrix[i].append(point.atlas_coordinate[0])
+ temp_matrix[i].append(point.atlas_coordinate[1])
+ temp_matrix[i].append(point.atlas_coordinate[2])
+ temp_matrix[i].append(point.structure)
+ temp_matrix[i].append(point.hemisphere)
+
+ df = pd.DataFrame(temp_matrix, columns=df.columns, index=None)
+ df.to_csv(all_points_filename, index=False)
+
+
+def get_region_totals(
+ points, structures_with_points, volume_csv_path, output_filename
+):
+ structures_with_points = list(structures_with_points)
+
+ point_numbers = pd.DataFrame(
+ columns=("structure_name", "hemisphere", "cell_count")
+ )
+ for structure in structures_with_points:
+ for hemisphere in ("left", "right"):
+ n_points = len(
+ [
+ point
+ for point in points
+ if point.structure == structure
+ and point.hemisphere == hemisphere
+ ]
+ )
+ if n_points:
+ point_numbers = point_numbers.append(
+ {
+ "structure_name": structure,
+ "hemisphere": hemisphere,
+ "cell_count": n_points,
+ },
+ ignore_index=True,
+ )
+ sorted_point_numbers = point_numbers.sort_values(
+ by=["cell_count"], ascending=False
+ )
+
+ combined_hemispheres = combine_df_hemispheres(sorted_point_numbers)
+ df = calculate_densities(combined_hemispheres, volume_csv_path)
+ df = sanitise_df(df)
+
+ df.to_csv(output_filename, index=False)
+
+
+def transform_points_to_downsampled_space(
+ points: np.ndarray,
+ target_space: bgs.AnatomicalSpace,
+ source_space: bgs.AnatomicalSpace,
+ output_filename: Optional[os.PathLike] = None,
+) -> np.ndarray:
+ """
+ Parameters
+ ----------
+ points :
+ Points in the original space.
+ target_space :
+ Target anatomical space.
+ source_space :
+ Source anatomical space of ``points``.
+ output_filename :
+ File to save downsampled points to. Points are saved as a HDF file
+ using pandas.
+
+ Returns
+ -------
+ downsampled_points
+ Points transformed to the downsampled space.
+ """
+ points = source_space.map_points_to(target_space, points)
+ if output_filename is not None:
+ df = pd.DataFrame(points)
+ df.to_hdf(output_filename, key="df", mode="w")
+ return points
+
+
+def transform_points_to_atlas_space(
+ points: np.ndarray,
+ source_space: bgs.AnatomicalSpace,
+ atlas: BrainGlobeAtlas,
+ deformation_field_paths: List[os.PathLike],
+ downsampled_space: bgs.AnatomicalSpace,
+ downsampled_points_path: Optional[os.PathLike] = None,
+ atlas_points_path: Optional[os.PathLike] = None,
+) -> np.ndarray:
+ """
+ Transform points to an atlas space.
+
+ The points are first downsampled to the atlas resolution, and then
+ transformed to the atlas space.
+ """
+ downsampled_points = transform_points_to_downsampled_space(
+ points,
+ downsampled_space,
+ source_space,
+ output_filename=downsampled_points_path,
+ )
+ return transform_points_downsampled_to_atlas_space(
+ downsampled_points,
+ atlas,
+ deformation_field_paths,
+ output_filename=atlas_points_path,
+ )
+
+
+def transform_points_downsampled_to_atlas_space(
+ downsampled_points: np.ndarray,
+ atlas: BrainGlobeAtlas,
+ deformation_field_paths: List[os.PathLike],
+ output_filename: Optional[os.PathLike] = None,
+) -> np.ndarray:
+ """
+ Parameters
+ ----------
+ downsampled_points :
+ Points already downsampled to the atlas resolution.
+ atlas :
+ Target atlas.
+ deformation_field_paths :
+ File paths to the deformation fields.
+ output_filename :
+ File to save transformed points to. Points are saved as a HDF file
+ using pandas.
+
+ Returns
+ -------
+ transformed_points
+ Points transformed to the atlas space.
+ """
+ field_scales = [int(1000 / resolution) for resolution in atlas.resolution]
+ points: List[List] = [[], [], []]
+ for axis, deformation_field_path in enumerate(deformation_field_paths):
+ deformation_field = tifffile.imread(deformation_field_path)
+ for point in downsampled_points:
+ point = [int(round(p)) for p in point]
+ points[axis].append(
+ int(
+ round(
+ field_scales[axis]
+ * deformation_field[point[0], point[1], point[2]]
+ )
+ )
+ )
+
+ transformed_points = np.array(points).T
+
+ if output_filename is not None:
+ df = pd.DataFrame(transformed_points)
+ df.to_hdf(output_filename, key="df", mode="w")
+
+ return transformed_points
+
+
+def run(args, cells, atlas, downsampled_space):
+ deformation_field_paths = [
+ args.brainreg_paths.deformation_field_0,
+ args.brainreg_paths.deformation_field_1,
+ args.brainreg_paths.deformation_field_2,
+ ]
+
+ cell_list = []
+ for cell in cells:
+ cell_list.append([cell.z, cell.y, cell.x])
+ cells = np.array(cell_list)
+
+ run_analysis(
+ cells,
+ args.signal_planes_paths[0],
+ args.orientation,
+ args.voxel_sizes,
+ atlas,
+ deformation_field_paths,
+ downsampled_space,
+ args.paths.downsampled_points,
+ args.paths.atlas_points,
+ args.paths.brainrender_points,
+ args.paths.abc4d_points,
+ args.brainreg_paths.volume_csv_path,
+ args.paths.all_points_csv,
+ args.paths.summary_csv,
+ )
+
+
+def run_analysis(
+ cells,
+ signal_planes,
+ orientation,
+ voxel_sizes,
+ atlas,
+ deformation_field_paths,
+ downsampled_space,
+ downsampled_points_path,
+ atlas_points_path,
+ brainrender_points_path,
+ abc4d_points_path,
+ volume_csv_path,
+ all_points_csv_path,
+ summary_csv_path,
+):
+ source_shape = tuple(
+ imio.get_size_image_from_file_paths(signal_planes).values()
+ )
+ source_shape = (source_shape[2], source_shape[1], source_shape[0])
+
+ source_space = bgs.AnatomicalSpace(
+ orientation,
+ shape=source_shape,
+ resolution=[float(i) for i in voxel_sizes],
+ )
+
+ transformed_cells = transform_points_to_atlas_space(
+ cells,
+ source_space,
+ atlas,
+ deformation_field_paths,
+ downsampled_space,
+ downsampled_points_path=downsampled_points_path,
+ atlas_points_path=atlas_points_path,
+ )
+
+ logging.info("Summarising cell positions")
+ points = summarise_points(
+ cells,
+ transformed_cells,
+ atlas,
+ volume_csv_path,
+ all_points_csv_path,
+ summary_csv_path,
+ )
+ logging.info("Exporting data")
+ export_points(
+ points,
+ transformed_cells,
+ atlas.resolution[0],
+ brainrender_points_path,
+ abc4d_points_path,
+ )
diff --git a/cellfinder/export/__init__.py b/cellfinder/export/__init__.py
new file mode 100644
index 00000000..e69de29b
diff --git a/cellfinder/export/abc4d.py b/cellfinder/export/abc4d.py
new file mode 100644
index 00000000..ff24f526
--- /dev/null
+++ b/cellfinder/export/abc4d.py
@@ -0,0 +1,20 @@
+import logging
+
+import numpy as np
+
+
+def export_points(
+ point_info,
+ resolution,
+ output_filename,
+):
+ logging.info("Exporting to abc4d")
+
+ point_arrays = []
+ for point in point_info:
+ point.atlas_coordinate = point.atlas_coordinate * resolution
+ point_arrays.append(
+ np.append(point.atlas_coordinate, point.structure_id)
+ )
+
+ np.save(output_filename, np.vstack(point_arrays).astype(np.float64))
diff --git a/cellfinder/export/brainrender.py b/cellfinder/export/brainrender.py
new file mode 100644
index 00000000..cb3421ba
--- /dev/null
+++ b/cellfinder/export/brainrender.py
@@ -0,0 +1,12 @@
+import logging
+
+import numpy as np
+
+
+def export_points(
+ points,
+ resolution,
+ output_filename,
+):
+ logging.info("Exporting to brainrender")
+ np.save(output_filename, points * resolution)
diff --git a/cellfinder/export/export.py b/cellfinder/export/export.py
new file mode 100644
index 00000000..1701f8a5
--- /dev/null
+++ b/cellfinder/export/export.py
@@ -0,0 +1,9 @@
+from cellfinder.export.abc4d import export_points as abc4d_export
+from cellfinder.export.brainrender import export_points as brainrender_export
+
+
+def export_points(
+ point_info, points, resolution, brainrender_points_path, abc4d_points_path
+):
+ brainrender_export(points, resolution, brainrender_points_path)
+ abc4d_export(point_info, resolution, abc4d_points_path)
diff --git a/cellfinder/extract/__init__.py b/cellfinder/extract/__init__.py
new file mode 100644
index 00000000..e69de29b
diff --git a/cellfinder/extract/extract_cubes.py b/cellfinder/extract/extract_cubes.py
new file mode 100644
index 00000000..54a0b8b3
--- /dev/null
+++ b/cellfinder/extract/extract_cubes.py
@@ -0,0 +1,492 @@
+"""
+Cube extraction for CNN-based classification.
+
+Based on, and mostly copied from,
+https://github.com/SainsburyWellcomeCentre/cell_count_analysis by
+Charly Rousseau (https://github.com/crousseau).
+"""
+
+import logging
+import os
+from collections import deque
+from concurrent.futures import ProcessPoolExecutor
+from datetime import datetime
+from math import floor
+
+import numpy as np
+from brainglobe_utils.cells.cells import group_cells_by_z
+from brainglobe_utils.general.numerical import is_even
+from brainglobe_utils.general.system import get_num_processes
+from numpy.linalg.linalg import LinAlgError
+from skimage import transform
+from tifffile import tifffile
+from tqdm import tqdm
+
+from cellfinder.tools import image_processing as img_tools
+from cellfinder.tools import system
+
+
+class StackSizeError(Exception):
+ pass
+
+
+class Cube(object):
+ class CubeBoundaryError(Exception):
+ pass
+
+ def __init__(
+ self,
+ cell,
+ channel,
+ stacks,
+ x_pix_um=1,
+ y_pix_um=1,
+ x_pix_um_network=1,
+ y_pix_um_network=1,
+ final_depth=20,
+ width=50,
+ height=50,
+ depth=20,
+ interpolation_order=0,
+ ):
+ self.cell = cell
+ self.channel = channel
+ self.stack = stacks[self.channel]
+ self.scale_cubes = False
+ self.scale_cubes_in_z = False
+ self.interpolation_order = interpolation_order
+ self.rescaling_factor_x = 1
+ self.rescaling_factor_y = 1
+
+ if x_pix_um != x_pix_um_network:
+ self.rescaling_factor_x = float(x_pix_um_network) / float(x_pix_um)
+ self.scale_cubes = True
+ if y_pix_um != y_pix_um_network:
+ self.rescaling_factor_y = float(y_pix_um_network) / float(y_pix_um)
+ self.scale_cubes = True
+
+ # This should be decided earlier
+ if depth != final_depth:
+ self.rescaling_factor_z = final_depth / depth
+ self.scale_cubes_in_z = True
+
+ self.width = width
+ self.height = height
+ self.depth = depth
+ self.final_depth = depth
+
+ self.empty = False
+ self.data = self.extract_cube(self.stack)
+
+ def __str__(self): # TODO: 0 pad (when changing classification)
+ prefix = "pCell"
+ return "{}z{}y{}x{}Ch{}.tif".format(
+ prefix, self.z, self.y, self.x, self.channel
+ )
+
+ @property
+ def x(self):
+ return self.cell.x
+
+ @property
+ def y(self):
+ return self.cell.y
+
+ @property
+ def z(self):
+ return self.cell.z
+
+ @property
+ def stack_x_max(self):
+ return self.stack.shape[1]
+
+ @property
+ def stack_y_max(self):
+ return self.stack.shape[0]
+
+ @property
+ def stack_z_max(self):
+ return self.stack.shape[2]
+
+ def extract_cube(self, stack):
+ """
+
+ :param np.array stack:
+ :return:
+ """
+ try:
+ self.check_boundaries()
+ except Cube.CubeBoundaryError:
+ return self.make_blank_data()
+
+ half_width, half_height, x0, x1, y0, y1 = self.get_boundaries()
+ data = stack[y0:y1, x0:x1, :]
+ if data.ndim < 3:
+ logging.error(
+ "Error occurred during extraction of cube {}, "
+ "original shape: {}, dtype: {}".format(
+ self, data.shape, data.dtype
+ )
+ )
+ return self.make_blank_data()
+
+ out = []
+ # TODO: combine these scalings
+ for i in range(self.depth):
+ if self.scale_cubes:
+ try:
+ # To match classification 1um rescaling
+ inv_rescale_x = 1 / self.rescaling_factor_x
+ inv_rescale_y = 1 / self.rescaling_factor_y
+ plane = transform.rescale(
+ data[:, :, i],
+ scale=(inv_rescale_y, inv_rescale_x),
+ order=self.interpolation_order,
+ preserve_range=True,
+ multichannel=False,
+ mode="constant",
+ anti_aliasing=False,
+ )
+ except LinAlgError as err:
+ logging.error(
+ f"Error occurred during rescale of cube {self}, "
+ f"original shape: {data[:, :, i].shape}, dtype: "
+ f"{data[:, :, i].dtype}; {err}."
+ )
+
+ return self.make_blank_data()
+ else:
+ plane = data[:, :, i]
+
+ plane = self.adjust_size(plane)
+
+ out.append(plane)
+ out = np.array(out, dtype=np.uint16)
+
+ if self.scale_cubes_in_z:
+ try:
+ # To match classification cube scaling in z
+ # TODO: presumably this will have rounding errors
+ out = transform.rescale(
+ out,
+ scale=(self.rescaling_factor_z, 1, 1),
+ order=self.interpolation_order,
+ preserve_range=True,
+ multichannel=False,
+ mode="constant",
+ anti_aliasing=False,
+ )
+ out = out.astype(np.uint16)
+ except LinAlgError as err:
+ logging.error(
+ f"Error occurred during z-rescale of cube {self}, "
+ f"from depth: {self.depth} to final depth "
+ f"{self.final_depth} ; {err}"
+ )
+ print.error(
+ f"Error occurred during z-rescale of cube {self}, "
+ f"from depth: {self.depth} to final depth "
+ f"{self.final_depth} ; {err}"
+ )
+ return self.make_blank_data()
+ return out
+
+ def adjust_size(self, plane):
+ plane = self.crop_to_correct_size(plane)
+ plane = self.pad_to_correct_size(plane)
+ return plane
+
+ def pad_to_correct_size(self, plane):
+ return img_tools.pad_center_2d(
+ plane, x_size=self.width, y_size=self.height
+ )
+
+ def crop_to_correct_size(self, plane):
+ return img_tools.crop_center_2d(
+ plane, crop_x=self.width, crop_y=self.height
+ )
+
+ def get_boundaries(self):
+ if self.scale_cubes:
+ half_width = int(round(self.width / 2 * self.rescaling_factor_x))
+ half_height = int(round(self.height / 2 * self.rescaling_factor_y))
+ else:
+ half_width = int(round(self.width / 2))
+ half_height = int(round(self.height / 2))
+
+ x0 = int(self.x - half_width)
+ x1 = int(self.x + half_width)
+ y0 = int(self.y - half_height)
+ y1 = int(self.y + half_height)
+ return half_width, half_height, x0, x1, y0, y1
+
+ def make_blank_data(self):
+ self.empty = True
+ return np.zeros((self.depth, self.width, self.height), dtype=np.uint16)
+
+ def check_boundaries(self):
+ out_bounds_message = (
+ "Cell candidate (x:{}, y:{}, z:{}) too close to the edge of the"
+ " image for cube extraction."
+ ).format(self.x, self.y, self.z)
+
+ if self.stack_z_max < self.depth:
+ error_msg = "Stack has {} planes but cube needs {}".format(
+ self.stack_z_max, self.depth
+ )
+ logging.error(error_msg)
+
+ raise Cube.CubeBoundaryError(error_msg)
+ boundaries = self.get_boundaries()
+ half_height, half_width, x0, x1, y0, y1 = boundaries
+ if x0 < 0 or y0 < 0:
+ logging.warning(out_bounds_message)
+ raise Cube.CubeBoundaryError(out_bounds_message)
+
+ # WARNING: dimensions inverted (numpy y=dim0, x=dim1)
+ elif x1 > self.stack_x_max or y1 > self.stack_y_max:
+ logging.warning(out_bounds_message)
+ raise Cube.CubeBoundaryError(out_bounds_message)
+
+
+def save_cubes(
+ cells,
+ planes_paths,
+ planes_to_read,
+ planes_shape,
+ voxel_sizes,
+ network_voxel_sizes,
+ num_planes_for_cube=20,
+ cube_width=50,
+ cube_height=50,
+ cube_depth=20,
+ thread_id=0,
+ output_dir="",
+ save_empty_cubes=False,
+):
+ """
+
+ :param cells:
+ :param planes_paths:
+ :param planes_to_read:
+ :param planes_shape:
+ :param x_pix_um:
+ :param y_pix_um:
+ :param x_pix_um_network:
+ :param y_pix_um_network:
+ :param num_planes_for_cube:
+ :param cube_width:
+ :param cube_height:
+ :param cube_depth:
+ :param thread_id:
+ :param output_dir:
+ :param save_empty_cubes:
+ :return:
+ """
+ channels = list(planes_paths.keys())
+ stack_shape = planes_shape + (num_planes_for_cube,)
+ stacks = {}
+ planes_queues = {}
+ for ch in channels:
+ stacks[ch] = np.zeros(stack_shape, dtype=np.uint16)
+ planes_queues[ch] = deque(maxlen=num_planes_for_cube)
+ for plane_idx in tqdm(planes_to_read, desc="Thread: {}".format(thread_id)):
+ for ch in channels:
+ plane_path = planes_paths[ch][plane_idx]
+ planes_queues[ch].append(tifffile.imread(plane_path))
+ if len(planes_queues[ch]) == num_planes_for_cube:
+ if is_even(num_planes_for_cube):
+ cell_z = int(plane_idx - num_planes_for_cube / 2 + 1)
+ else:
+ cell_z = int(
+ plane_idx - floor(num_planes_for_cube) / 2 + 1
+ )
+
+ for j, plane in enumerate(planes_queues[ch]):
+ stacks[ch][:, :, j] = plane
+
+ # ensures no cube_depth planes at the end
+ planes_queues[ch].popleft()
+ # required since we provide all cells
+ # TODO: if len(planes_queues[ch])
+ # < num_planes_for_cube -1: break
+ for cell in cells[cell_z]:
+ cube = Cube(
+ cell,
+ ch,
+ stacks,
+ x_pix_um=voxel_sizes[2],
+ y_pix_um=voxel_sizes[1],
+ x_pix_um_network=network_voxel_sizes[2],
+ y_pix_um_network=network_voxel_sizes[1],
+ final_depth=cube_depth,
+ width=cube_width,
+ height=cube_height,
+ depth=num_planes_for_cube,
+ )
+ if not cube.empty or (cube.empty and save_empty_cubes):
+ tifffile.imsave(
+ os.path.join(output_dir, str(cube)), cube.data
+ )
+
+
+def get_ram_requirement_per_process(
+ single_image_path, cube_depth, num_channels=2, copies=2
+):
+ """
+ Calculates how much RAM is needed per CPU core for cube extraction. Used
+ later to set the number of CPU cores to be used.
+
+ RAM requirement is currently:
+ image_size x n_cores x n_channels x cube_depth x copies
+
+ :param single_image_path: A single 2D image, used to find the size.
+ :param num_channels: Number of channels to be extracted
+ :param cube_depth: Depth of the cube to be extracted (z-planes)
+ to use.
+ :param int copies: How many copies of the data are loaded at any time
+ :return int: memory requirement for one CPU core
+ """
+
+ logging.debug(
+ "Determining how much RAM is needed for each CPU for cube extraction"
+ )
+ file_size = os.path.getsize(single_image_path)
+ total_mem_need = file_size * num_channels * cube_depth * copies
+
+ logging.debug(
+ "File size: {:.2f} GB, "
+ "number of channels: {}, "
+ "cube depth: {}, "
+ "Therefore theoretically {:.2f} GB is needed per CPU core."
+ "".format(
+ (file_size / (1024**3)),
+ num_channels,
+ cube_depth,
+ (total_mem_need / (1024**3)),
+ )
+ )
+
+ return total_mem_need
+
+
+def main(
+ cells,
+ cubes_output_dir,
+ planes_paths,
+ cube_depth,
+ cube_width,
+ cube_height,
+ voxel_sizes,
+ network_voxel_sizes,
+ max_ram,
+ n_free_cpus=4,
+ save_empty_cubes=False,
+):
+ start_time = datetime.now()
+
+ if voxel_sizes[0] != network_voxel_sizes[0]:
+ plane_scaling_factor = float(network_voxel_sizes[0]) / float(
+ voxel_sizes[0]
+ )
+ num_planes_needed_for_cube = round(cube_depth * plane_scaling_factor)
+ else:
+ num_planes_needed_for_cube = cube_depth
+
+ if num_planes_needed_for_cube > len(planes_paths[0]):
+ raise StackSizeError(
+ "The number of planes provided is not sufficient "
+ "for any cubes to be extracted. Please check the "
+ "input data"
+ )
+
+ first_plane = tifffile.imread(list(planes_paths.values())[0][0])
+
+ planes_shape = first_plane.shape
+ brain_depth = len(list(planes_paths.values())[0])
+
+ # TODO: use to assert all centre planes processed
+ center_planes = sorted(list(set([cell.z for cell in cells])))
+
+ # REFACTOR: rename (clashes with different meaning of planes_to_read below)
+ planes_to_read = np.zeros(brain_depth, dtype=bool)
+
+ if is_even(num_planes_needed_for_cube):
+ half_nz = num_planes_needed_for_cube // 2
+ # WARNING: not centered because even
+ for p in center_planes:
+ planes_to_read[p - half_nz : p + half_nz] = 1
+ else:
+ half_nz = num_planes_needed_for_cube // 2
+ # centered
+ for p in center_planes:
+ planes_to_read[p - half_nz : p + half_nz + 1] = 1
+
+ planes_to_read = np.where(planes_to_read)[0]
+
+ if not planes_to_read.size:
+ logging.error(
+ f"No planes found, you need at the very least "
+ f"{num_planes_needed_for_cube} "
+ f"planes to proceed (i.e. cube z size)"
+ f"Brain z dimension is {brain_depth}.",
+ stack_info=True,
+ )
+ raise ValueError(
+ f"No planes found, you need at the very least "
+ f"{num_planes_needed_for_cube} "
+ f"planes to proceed (i.e. cube z size)"
+ f"Brain z dimension is {brain_depth}."
+ )
+ # TODO: check if needs to flip args.cube_width and args.cube_height
+ cells_groups = group_cells_by_z(cells)
+
+ # copies=2 is set because at all times there is a plane queue (deque)
+ # and an array passed to `Cube`
+ ram_per_process = get_ram_requirement_per_process(
+ planes_paths[0][0],
+ num_planes_needed_for_cube,
+ copies=2,
+ )
+ n_processes = get_num_processes(
+ min_free_cpu_cores=n_free_cpus,
+ ram_needed_per_process=ram_per_process,
+ n_max_processes=len(planes_to_read),
+ fraction_free_ram=0.2,
+ max_ram_usage=system.memory_in_bytes(max_ram, "GB"),
+ )
+ # TODO: don't need to extract cubes from all channels if
+ # n_signal_channels>1
+ with ProcessPoolExecutor(max_workers=n_processes) as executor:
+ n_planes_per_chunk = len(planes_to_read) // n_processes
+ for i in range(n_processes):
+ start_idx = i * n_planes_per_chunk
+ end_idx = (
+ start_idx + n_planes_per_chunk + num_planes_needed_for_cube - 1
+ )
+ if end_idx > planes_to_read[-1]:
+ end_idx = None
+ sub_planes_to_read = planes_to_read[start_idx:end_idx]
+
+ executor.submit(
+ save_cubes,
+ cells_groups,
+ planes_paths,
+ sub_planes_to_read,
+ planes_shape,
+ voxel_sizes,
+ network_voxel_sizes,
+ num_planes_for_cube=num_planes_needed_for_cube,
+ cube_width=cube_width,
+ cube_height=cube_height,
+ cube_depth=cube_depth,
+ thread_id=i,
+ output_dir=cubes_output_dir,
+ save_empty_cubes=save_empty_cubes,
+ )
+
+ total_cubes = system.get_number_of_files_in_dir(cubes_output_dir)
+ time_taken = datetime.now() - start_time
+ logging.info(
+ "All cubes ({}) extracted in: {}".format(total_cubes, time_taken)
+ )
diff --git a/cellfinder/figures/__init__.py b/cellfinder/figures/__init__.py
new file mode 100644
index 00000000..e69de29b
diff --git a/cellfinder/figures/figures.py b/cellfinder/figures/figures.py
new file mode 100644
index 00000000..59380542
--- /dev/null
+++ b/cellfinder/figures/figures.py
@@ -0,0 +1,16 @@
+import logging
+
+from cellfinder.figures import heatmap
+
+
+def run(args, atlas, downsampled_shape):
+ logging.info("Generating heatmap")
+ heatmap.run(
+ args.paths.downsampled_points,
+ atlas,
+ downsampled_shape,
+ args.brainreg_paths.registered_atlas,
+ args.paths.heatmap,
+ smoothing=args.heatmap_smooth,
+ mask=args.mask_figures,
+ )
diff --git a/cellfinder/figures/heatmap.py b/cellfinder/figures/heatmap.py
new file mode 100644
index 00000000..18af4a09
--- /dev/null
+++ b/cellfinder/figures/heatmap.py
@@ -0,0 +1,62 @@
+import logging
+from pathlib import Path
+
+import imio
+import numpy as np
+import pandas as pd
+import tifffile
+from brainglobe_utils.general.system import ensure_directory_exists
+from brainglobe_utils.image.masking import mask_image_threshold
+from brainglobe_utils.image.scale import scale_and_convert_to_16_bits
+from skimage.filters import gaussian
+
+
+def get_bins(image_size, bin_sizes):
+ """
+ Given an image size, and bin size, return a list of the bin boundaries
+ :param image_size: Size of the final image (tuple/list)
+ :param bin_sizes: Bin sizes corresponding to the dimensions of
+ "image_size" (tuple/list)
+ :return: List of arrays of bin boundaries
+ """
+ bins = []
+ for dim in range(0, len(image_size)):
+ bins.append(np.arange(0, image_size[dim] + 1, bin_sizes[dim]))
+ return bins
+
+
+def run(
+ downsampled_points,
+ atlas,
+ downsampled_shape,
+ registered_atlas_path,
+ output_filename,
+ smoothing=None,
+ mask=True,
+):
+ points = pd.read_hdf(downsampled_points).values
+
+ bins = get_bins(downsampled_shape, (1, 1, 1))
+ heatmap_array, _ = np.histogramdd(points, bins=bins)
+ heatmap_array = heatmap_array.astype(np.uint16)
+
+ if smoothing is not None:
+ logging.debug("Smoothing heatmap")
+ # assume isotropic atlas
+ smoothing = int(round(smoothing / atlas.resolution[0]))
+ heatmap_array = gaussian(heatmap_array, sigma=smoothing)
+
+ if mask:
+ logging.debug("Masking image based on registered atlas")
+ atlas = tifffile.imread(registered_atlas_path)
+ heatmap_array = mask_image_threshold(heatmap_array, atlas)
+ del atlas
+
+ logging.debug("Saving heatmap")
+ heatmap_array = scale_and_convert_to_16_bits(heatmap_array)
+
+ logging.debug("Ensuring output directory exists")
+ ensure_directory_exists(Path(output_filename).parent)
+
+ logging.debug("Saving heatmap image")
+ imio.to_tiff(heatmap_array, output_filename)
diff --git a/cellfinder/main.py b/cellfinder/main.py
new file mode 100644
index 00000000..42046e39
--- /dev/null
+++ b/cellfinder/main.py
@@ -0,0 +1,232 @@
+"""
+main
+===============
+
+Runs each part of the cellfinder pipeline in turn.
+
+N.B imports are within functions to prevent tensorflow being imported before
+it's warnings are silenced
+"""
+
+import logging
+import os
+from datetime import datetime
+
+import bg_space as bgs
+import tifffile
+from brainglobe_utils.cells.cells import MissingCellsError
+from brainglobe_utils.general.system import ensure_directory_exists
+from brainglobe_utils.IO.cells import get_cells, save_cells
+from cellfinder_core.main import suppress_tf_logging, tf_suppress_log_messages
+
+BRAINREG_PRE_PROCESSING_ARGS = None
+
+
+def get_downsampled_space(atlas, downsampled_image_path):
+ target_shape = tifffile.imread(downsampled_image_path).shape
+ downsampled_space = bgs.AnatomicalSpace(
+ atlas.metadata["orientation"],
+ shape=target_shape,
+ resolution=atlas.resolution,
+ )
+ return downsampled_space
+
+
+def cells_exist(points_file):
+ try:
+ get_cells(points_file, cells_only=True)
+ return True
+ except MissingCellsError:
+ return False
+
+
+def main():
+ suppress_tf_logging(tf_suppress_log_messages)
+ from brainreg.main import main as register
+
+ from cellfinder.tools import prep
+
+ start_time = datetime.now()
+ args, arg_groups, what_to_run, atlas = prep.prep_cellfinder_general()
+
+ if what_to_run.register:
+ # TODO: add register_part_brain option
+ logging.info("Registering to atlas")
+ args, additional_images_downsample = prep.prep_registration(args)
+ register(
+ args.atlas,
+ args.orientation,
+ args.target_brain_path,
+ args.brainreg_paths,
+ args.voxel_sizes,
+ arg_groups["NiftyReg registration backend options"],
+ BRAINREG_PRE_PROCESSING_ARGS,
+ sort_input_file=args.sort_input_file,
+ n_free_cpus=args.n_free_cpus,
+ additional_images_downsample=additional_images_downsample,
+ backend=args.backend,
+ debug=args.debug,
+ )
+
+ else:
+ logging.info("Skipping registration")
+
+ if len(args.signal_planes_paths) > 1:
+ base_directory = args.output_dir
+
+ for idx, signal_paths in enumerate(args.signal_planes_paths):
+ channel = args.signal_ch_ids[idx]
+ logging.info("Processing channel: " + str(channel))
+ channel_directory = os.path.join(
+ base_directory, "channel_" + str(channel)
+ )
+ if not os.path.exists(channel_directory):
+ os.makedirs(channel_directory)
+
+ # prep signal channel specific args
+ args.signal_planes_paths[0] = signal_paths
+ # TODO: don't overwrite args.output_dir - use Paths instead
+ args.output_dir = channel_directory
+ args.signal_channel = channel
+ # Run for each channel
+ run_all(args, what_to_run, atlas)
+
+ else:
+ args.signal_channel = args.signal_ch_ids[0]
+ run_all(args, what_to_run, atlas)
+ logging.info(
+ "Finished. Total time taken: {}".format(datetime.now() - start_time)
+ )
+
+
+def run_all(args, what_to_run, atlas):
+ from cellfinder_core.classify import classify
+ from cellfinder_core.detect import detect
+ from cellfinder_core.tools import prep
+ from cellfinder_core.tools.IO import read_with_dask
+
+ from cellfinder.analyse import analyse
+ from cellfinder.figures import figures
+ from cellfinder.tools.prep import (
+ prep_candidate_detection,
+ prep_channel_specific_general,
+ )
+
+ points = None
+ signal_array = None
+ args, what_to_run = prep_channel_specific_general(args, what_to_run)
+
+ if what_to_run.detect:
+ logging.info("Detecting cell candidates")
+ args = prep_candidate_detection(args)
+ signal_array = read_with_dask(
+ args.signal_planes_paths[args.signal_channel]
+ )
+
+ points = detect.main(
+ signal_array,
+ args.start_plane,
+ args.end_plane,
+ args.voxel_sizes,
+ args.soma_diameter,
+ args.max_cluster_size,
+ args.ball_xy_size,
+ args.ball_z_size,
+ args.ball_overlap_fraction,
+ args.soma_spread_factor,
+ args.n_free_cpus,
+ args.log_sigma_size,
+ args.n_sds_above_mean_thresh,
+ save_planes=args.save_planes,
+ plane_directory=args.plane_directory,
+ )
+ ensure_directory_exists(args.paths.points_directory)
+
+ save_cells(
+ points,
+ args.paths.detected_points,
+ save_csv=args.save_csv,
+ artifact_keep=args.artifact_keep,
+ )
+
+ else:
+ logging.info("Skipping cell detection")
+ points = get_cells(args.paths.detected_points)
+
+ if what_to_run.classify:
+ model_weights = prep.prep_classification(
+ args.trained_model,
+ args.model_weights,
+ args.install_path,
+ args.model,
+ args.n_free_cpus,
+ )
+ if what_to_run.classify:
+ if points is None:
+ points = get_cells(args.paths.detected_points)
+ if signal_array is None:
+ signal_array = read_with_dask(
+ args.signal_planes_paths[args.signal_channel]
+ )
+ logging.info("Running cell classification")
+ background_array = read_with_dask(args.background_planes_path[0])
+
+ points = classify.main(
+ points,
+ signal_array,
+ background_array,
+ args.n_free_cpus,
+ args.voxel_sizes,
+ args.network_voxel_sizes,
+ args.batch_size,
+ args.cube_height,
+ args.cube_width,
+ args.cube_depth,
+ args.trained_model,
+ model_weights,
+ args.network_depth,
+ )
+ save_cells(
+ points,
+ args.paths.classified_points,
+ save_csv=args.save_csv,
+ )
+
+ what_to_run.cells_exist = cells_exist(args.paths.classified_points)
+
+ else:
+ logging.info("No cells were detected, skipping classification.")
+
+ else:
+ logging.info("Skipping cell classification")
+
+ what_to_run.update_if_cells_required()
+
+ if what_to_run.analyse or what_to_run.figures:
+ downsampled_space = get_downsampled_space(
+ atlas, args.brainreg_paths.boundaries_file_path
+ )
+
+ if what_to_run.analyse:
+ points = get_cells(args.paths.classified_points, cells_only=True)
+ if len(points) == 0:
+ logging.info("No cells detected, skipping cell position analysis")
+ else:
+ logging.info("Analysing cell positions")
+ analyse.run(args, points, atlas, downsampled_space)
+ else:
+ logging.info("Skipping cell position analysis")
+
+ if what_to_run.figures:
+ points = get_cells(args.paths.classified_points, cells_only=True)
+ if len(points) == 0:
+ logging.info("No cells detected, skipping")
+ else:
+ logging.info("Generating figures")
+ figures.run(args, atlas, downsampled_space.shape)
+ else:
+ logging.info("Skipping figure generation")
+
+
+if __name__ == "__main__":
+ main()
diff --git a/cellfinder/tools/__init__.py b/cellfinder/tools/__init__.py
new file mode 100644
index 00000000..e69de29b
diff --git a/cellfinder/tools/image_processing.py b/cellfinder/tools/image_processing.py
new file mode 100644
index 00000000..742220d6
--- /dev/null
+++ b/cellfinder/tools/image_processing.py
@@ -0,0 +1,87 @@
+import numpy as np
+from brainglobe_utils.general.numerical import is_even
+
+
+def crop_center_2d(img, crop_x=None, crop_y=None):
+ """
+ Crops the centre of a 2D image, and returns a smaller array. If the desired
+ dimension is larger than the original dimension, nothing is changed.
+ :param img: 2D input image
+ :param crop_x: New length in x (default: None, which does nothing)
+ :param crop_y: New length in y (default: None, which does nothing)
+ :return: New, smaller array
+ """
+
+ y, x = img.shape
+
+ # TODO: simplify
+ if crop_x is not None:
+ if crop_x >= x:
+ start_x = 0
+ crop_x = x
+ else:
+ start_x = x // 2 - (crop_x // 2)
+ else:
+ start_x = 0
+ crop_x = x
+
+ if crop_y is not None:
+ if crop_y >= y:
+ start_y = 0
+ crop_y = y
+ else:
+ start_y = y // 2 - (crop_y // 2)
+ else:
+ start_y = 0
+ crop_y = y
+
+ return img[start_y : start_y + crop_y, start_x : start_x + crop_x]
+
+
+def pad_center_2d(img, x_size=None, y_size=None, pad_mode="edge"):
+ """
+ Pads the edges of a 2D image, and returns a larger image. If the desired
+ dimension is smaller than the original dimension, nothing is changed.
+ :param img: 2D input image
+ :param x_size: New length in x (default: None, which does nothing)
+ :param y_size: New length in y (default: None, which does nothing)
+ :return: New, larger array
+ """
+
+ y, x = img.shape
+
+ # TODO: simplify
+
+ if x_size is None:
+ x_pad = 0
+ elif x_size <= x:
+ x_pad = 0
+ else:
+ x_pad = x_size - x
+
+ if y_size is None:
+ y_pad = 0
+ elif y_size <= y:
+ y_pad = 0
+ else:
+ y_pad = y_size - y
+
+ if x_pad > 0:
+ if is_even(x_pad):
+ x_front = x_back = int(x_pad / 2)
+ else:
+ x_front = int(x_pad // 2)
+ x_back = int(x_front + 1)
+ else:
+ x_front = x_back = 0
+
+ if y_pad > 0:
+ if is_even(y_pad):
+ y_front = y_back = int(y_pad / 2)
+ else:
+ y_front = int(y_pad // 2)
+ y_back = int(y_front + 1)
+ else:
+ y_front = y_back = 0
+
+ return np.pad(img, ((y_front, y_back), (x_front, x_back)), pad_mode)
diff --git a/cellfinder/tools/parser.py b/cellfinder/tools/parser.py
new file mode 100644
index 00000000..e44f9162
--- /dev/null
+++ b/cellfinder/tools/parser.py
@@ -0,0 +1,460 @@
+"""
+parser
+========
+All the various arguments that may be needed by the various submodules.
+Defined together so they can also be called by any other entry points.
+"""
+
+
+from argparse import (
+ ArgumentDefaultsHelpFormatter,
+ ArgumentParser,
+ ArgumentTypeError,
+)
+
+from brainglobe_utils.general.numerical import (
+ check_positive_float,
+ check_positive_int,
+)
+from brainreg.cli import atlas_parse, geometry_parser, niftyreg_parse
+from brainreg.cli import backend_parse as brainreg_backend_parse
+from cellfinder_core.download.cli import (
+ download_directory_parser,
+ model_parser,
+)
+from cellfinder_core.tools.source_files import source_custom_config_cellfinder
+
+from cellfinder import __version__
+
+# TODO: Gradually move all paths as strings to Path objects
+
+models = {
+ "18": "18-layer",
+ "34": "34-layer",
+ "50": "50-layer",
+ "101": "101-layer",
+ "152": "152-layer",
+}
+
+
+def valid_model_depth(depth):
+ """
+ Ensures a correct existing_model is chosen
+ :param value: Input value
+ :param models: Dict of allowed models
+ :return: Input value, if it corresponds to a valid existing_model
+ """
+
+ if depth in models.keys():
+ return depth
+ else:
+ raise ArgumentTypeError(
+ f"Model depth: {depth} is not valid. Please "
+ f"choose one of: {list(models.keys())}"
+ )
+
+
+def cellfinder_parser():
+ parser = ArgumentParser(formatter_class=ArgumentDefaultsHelpFormatter)
+ parser = main_parse(parser)
+ parser = config_parse(parser)
+ parser = pixel_parser(parser)
+ parser = run_parse(parser)
+ parser = io_parse(parser)
+ parser = cell_detect_parse(parser)
+ parser = classification_parse(parser)
+ parser = cube_extract_parse(parser)
+ parser = misc_parse(parser)
+ parser = model_parser(parser)
+ parser = figures_parse(parser)
+ parser = download_directory_parser(parser)
+
+ # brainreg options
+ parser = atlas_parse(parser)
+ parser = geometry_parser(parser)
+ parser = brainreg_backend_parse(parser)
+ # This needs to be abstracted away into brainreg for multiple backends
+ parser = niftyreg_parse(parser)
+
+ return parser
+
+
+def main_parse(parser):
+ main_parser = parser.add_argument_group("General options")
+
+ main_parser.add_argument(
+ "-s",
+ "--signal-planes-paths",
+ dest="signal_planes_paths",
+ type=str,
+ nargs="+",
+ required=True,
+ help="Path to the directory of the signal files. Can also be a text"
+ "file pointing to the files.",
+ )
+ main_parser.add_argument(
+ "-b",
+ "--background-planes-path",
+ dest="background_planes_path",
+ type=str,
+ nargs=1,
+ required=True,
+ help="Path to the directory of the background files. Can also be a "
+ "text file pointing to the files.",
+ )
+ main_parser.add_argument(
+ "-o",
+ "--output-dir",
+ dest="output_dir",
+ type=str,
+ required=True,
+ help="Output directory for all intermediate and final results.",
+ )
+ main_parser.add_argument(
+ "--signal-channel-ids",
+ dest="signal_ch_ids",
+ type=check_positive_int,
+ nargs="+",
+ help="Channel ID numbers, in the same order as 'signal-planes-paths'."
+ " Will default to '0, 1, 2' etc, but maybe useful to specify.",
+ )
+ main_parser.add_argument(
+ "--background-channel-id",
+ dest="background_ch_id",
+ type=check_positive_int,
+ help="Channel ID number, corresponding to 'background-planes-path'.",
+ )
+ main_parser.add_argument(
+ "--version",
+ action="version",
+ version="%(prog)s {version}".format(version=__version__),
+ )
+ return parser
+
+
+def pixel_parser(parser):
+ # TODO: separate the two groups. Image pixel sizes are needed in lots of
+ # places that the network pixel sizes are not
+ pixel_opt_parser = parser.add_argument_group(
+ "Options to define pixel sizes of raw data"
+ )
+ pixel_opt_parser.add_argument(
+ "-v",
+ "--voxel-sizes",
+ dest="voxel_sizes",
+ required=True,
+ nargs="+",
+ help="Voxel sizes in microns, in the order of data orientation. "
+ "e.g. '5 2 2'",
+ )
+ pixel_opt_parser.add_argument(
+ "--network-voxel-sizes",
+ dest="network_voxel_sizes",
+ nargs="+",
+ default=[5, 1, 1],
+ help="Voxel sizes in microns that the machine learning network was "
+ "trained on, in the order of data orientation. e.g. '5 2 2'."
+ "Set this to adjust the pixel sizes of the extracted cubes",
+ )
+ return parser
+
+
+def run_parse(parser):
+ run_parser = parser.add_argument_group(
+ "Options to disable part of cellfinder"
+ )
+ run_parser.add_argument(
+ "--no-detection",
+ dest="no_detection",
+ action="store_true",
+ help="Dont run cell candidate detection",
+ )
+
+ run_parser.add_argument(
+ "--no-classification",
+ dest="no_classification",
+ action="store_true",
+ help="Do not run cell classification",
+ )
+ run_parser.add_argument(
+ "--no-register",
+ dest="no_register",
+ action="store_true",
+ help="Do not perform registration",
+ )
+ run_parser.add_argument(
+ "--no-analyse",
+ dest="no_analyse",
+ action="store_true",
+ help="Do not analyse and export cell positions",
+ )
+ run_parser.add_argument(
+ "--no-figures",
+ dest="no_figures",
+ action="store_true",
+ help="Do not create figures (e.g. heatmap)",
+ )
+
+ return parser
+
+
+def io_parse(parser):
+ io_parser = parser.add_argument_group("Input & output options")
+ io_parser.add_argument(
+ "--start-plane",
+ dest="start_plane",
+ type=check_positive_int,
+ default=0,
+ help="The first plane to process in the Z dimension.",
+ )
+ io_parser.add_argument(
+ "--end-plane",
+ dest="end_plane",
+ type=int,
+ default=-1,
+ help="The last plane to process in the Z dimension.",
+ )
+ return parser
+
+
+def cell_detect_parse(parser):
+ # TODO: improve the help on these files
+ cell_detect_parser = parser.add_argument_group(
+ "Detection options", description="Cell detection options"
+ )
+ cell_detect_parser.add_argument(
+ "--save-planes",
+ dest="save_planes",
+ action="store_true",
+ help="Whether to save the individual planes after "
+ "processing and thresholding. Useful for debugging.",
+ )
+
+ cell_detect_parser.add_argument(
+ "--outlier-keep",
+ dest="outlier_keep",
+ action="store_true",
+ help="Dont remove putative cells that fall outside initial clusters",
+ )
+
+ cell_detect_parser.add_argument(
+ "--artifact-keep",
+ dest="artifact_keep",
+ action="store_true",
+ help="Save artifacts into the initial xml file",
+ )
+
+ cell_detect_parser.add_argument(
+ "--max-cluster-size",
+ dest="max_cluster_size",
+ type=check_positive_int,
+ default=100000,
+ help="Largest putative cell cluster (in cubic um) where "
+ "splitting should be attempted",
+ )
+
+ cell_detect_parser.add_argument(
+ "--soma-diameter",
+ dest="soma_diameter",
+ type=check_positive_float,
+ default=16,
+ help="The expected soma size in um in the x/y dimensions",
+ )
+
+ cell_detect_parser.add_argument(
+ "--ball-xy-size",
+ dest="ball_xy_size",
+ type=check_positive_int,
+ default=6,
+ help="The size in um of the ball used "
+ "for the morphological filter in the x/y dimensions",
+ )
+ cell_detect_parser.add_argument(
+ "--ball-z-size",
+ dest="ball_z_size",
+ type=check_positive_int,
+ default=15,
+ help="The size in um of the ball used "
+ "for the morphological filter in the z dimension",
+ )
+ cell_detect_parser.add_argument(
+ "--ball-overlap-fraction",
+ dest="ball_overlap_fraction",
+ type=check_positive_float,
+ default=0.6,
+ help="The fraction of the ball that has to cover thresholded pixels "
+ "for the centre pixel to be considered a nucleus pixel",
+ )
+
+ cell_detect_parser.add_argument(
+ "--log-sigma-size",
+ dest="log_sigma_size",
+ type=check_positive_float,
+ default=0.2,
+ help="The filter size used in the Laplacian of Gaussian filter to "
+ "enhance the cell intensities. Given as a fraction of the "
+ "soma-diameter.",
+ )
+ cell_detect_parser.add_argument(
+ "--threshold",
+ dest="n_sds_above_mean_thresh",
+ type=check_positive_float,
+ default=10,
+ help="The cell threshold, in multiples of the standard deviation"
+ "above the mean",
+ )
+ cell_detect_parser.add_argument(
+ "--soma-spread-factor",
+ dest="soma_spread_factor",
+ type=check_positive_float,
+ default=1.4,
+ help="Soma size spread factor (for splitting up cell clusters)",
+ )
+
+ return parser
+
+
+def classification_parse(parser):
+ classification_parser = parser.add_argument_group(
+ "Cell classification options"
+ )
+ classification_parser.add_argument(
+ "--trained-model",
+ dest="trained_model",
+ type=str,
+ help="Path to the trained model",
+ )
+ classification_parser.add_argument(
+ "--model-weights",
+ dest="model_weights",
+ type=str,
+ help="Path to existing model weights",
+ )
+ classification_parser.add_argument(
+ "--network-depth",
+ dest="network_depth",
+ type=valid_model_depth,
+ default="50",
+ help="Resnet depth (based on He et al. (2015)",
+ )
+ classification_parser.add_argument(
+ "--batch-size",
+ dest="batch_size",
+ type=check_positive_int,
+ default=32,
+ help="Batch size for classification. Can be adjusted depending on "
+ "GPU memory.",
+ )
+ return parser
+
+
+def cube_extract_parse(parser):
+ cube_extract_parser = parser.add_argument_group("Cube extraction options")
+ cube_extract_parser.add_argument(
+ "--cube-width",
+ dest="cube_width",
+ type=check_positive_int,
+ default=50,
+ help="The width of the cubes to extract (must be even)",
+ )
+ cube_extract_parser.add_argument(
+ "--cube-height",
+ dest="cube_height",
+ type=check_positive_int,
+ default=50,
+ help="The height of the cubes to extract (must be even)",
+ )
+ cube_extract_parser.add_argument(
+ "--cube-depth",
+ dest="cube_depth",
+ type=check_positive_int,
+ default=20,
+ help="The depth (z) of the cubes to extract",
+ )
+ cube_extract_parser.add_argument(
+ "--save-empty-cubes",
+ dest="save_empty_cubes",
+ action="store_true",
+ help="If a cube cannot be extracted (e.g. to close to the edge of"
+ "the image), save an empty cube instead. Useful to keep track"
+ "of all cell candidates.",
+ )
+ return parser
+
+
+def config_parse(parser):
+ config_opt_parser = parser.add_argument_group("Config options")
+ config_opt_parser.add_argument(
+ "--config",
+ dest="registration_config",
+ type=str,
+ default=source_custom_config_cellfinder(),
+ help="To supply your own, custom configuration file.",
+ )
+
+ return parser
+
+
+def figures_parse(parser):
+ figure_parser = parser.add_argument_group(
+ "Figure generation specific parameters"
+ )
+ figure_parser.add_argument(
+ "--heatmap-smoothing",
+ dest="heatmap_smooth",
+ type=check_positive_float,
+ default=100,
+ help="Gaussian smoothing sigma, in um.",
+ )
+ figure_parser.add_argument(
+ "--no-mask-figs",
+ dest="mask_figures",
+ action="store_false",
+ help="Don't mask the figures (removing any areas outside the brain,"
+ "from e.g. smoothing)",
+ )
+ return parser
+
+
+def misc_parse(parser):
+ misc_parser = parser.add_argument_group("Misc options")
+ misc_parser.add_argument(
+ "--n-free-cpus",
+ dest="n_free_cpus",
+ type=check_positive_int,
+ default=2,
+ help="The number of CPU cores on the machine to leave "
+ "unused by the program to spare resources.",
+ )
+ misc_parser.add_argument(
+ "--max-ram",
+ dest="max_ram",
+ type=check_positive_float,
+ default=None,
+ help="Maximum amount of RAM to use (in GB) - not currently fully "
+ "implemented for all parts of cellfinder",
+ )
+ misc_parser.add_argument(
+ "--save-csv",
+ dest="save_csv",
+ action="store_true",
+ help="Save .csv files of cell locations (in addition to xml)."
+ "Useful for importing into other software.",
+ )
+ misc_parser.add_argument(
+ "--debug",
+ dest="debug",
+ action="store_true",
+ help="Debug mode. Will increase verbosity of logging and save all "
+ "intermediate files for diagnosis of software issues.",
+ )
+ misc_parser.add_argument(
+ "--sort-input-file",
+ dest="sort_input_file",
+ action="store_true",
+ help="If set to true, the input text file will be sorted using "
+ "natural sorting. This means that the file paths will be "
+ "sorted as would be expected by a human and "
+ "not purely alphabetically",
+ )
+ return parser
diff --git a/cellfinder/tools/prep.py b/cellfinder/tools/prep.py
new file mode 100644
index 00000000..f52b5368
--- /dev/null
+++ b/cellfinder/tools/prep.py
@@ -0,0 +1,331 @@
+"""
+prep
+==================
+Functions to prepare files and directories needed for other functions
+"""
+
+
+import json
+import logging
+import os
+from argparse import Namespace
+from pathlib import PurePath
+
+from bg_atlasapi import BrainGlobeAtlas
+from brainglobe_utils.general.exceptions import CommandLineInputError
+from brainglobe_utils.general.system import ensure_directory_exists
+from brainreg.paths import Paths as BrainRegPaths
+from fancylog import fancylog
+
+import cellfinder as program_for_log
+import cellfinder.tools.parser as parser
+from cellfinder.tools import system, tools
+from cellfinder.tools.parser import cellfinder_parser
+
+
+def get_arg_groups(args, parser):
+ arg_groups = {}
+ for group in parser._action_groups:
+ group_dict = {
+ a.dest: getattr(args, a.dest, None) for a in group._group_actions
+ }
+ arg_groups[group.title] = Namespace(**group_dict)
+
+ return arg_groups
+
+
+def check_input_arg_existance(args):
+ """
+ Does a simple check to ensure that input files/paths exist. Prevents a typo
+ from causing cellfinder to only run partway. Doesn't check for validity
+ etc, just existance.
+ :param args: Cellfinder input arguments
+ """
+ check_list = [args.background_planes_path[0]]
+ check_list = check_list + args.signal_planes_paths
+
+ for path in check_list:
+ if path is not None:
+ system.catch_input_file_error(path)
+
+
+class Paths:
+ """
+ A single class to hold all file paths that cellfinder may need. Any paths
+ prefixed with "tmp__" refer to internal intermediate steps, and will be
+ deleted if "--debug" is not used.
+ """
+
+ def __init__(self, output_dir):
+ self.output_dir = output_dir
+ self.registration_output_folder = os.path.join(
+ self.output_dir, "registration"
+ )
+ self.metadata_path = os.path.join(self.output_dir, "cellfinder.json")
+ self.registration_metadata_path = os.path.join(
+ self.registration_output_folder, "brainreg.json"
+ )
+
+ def make_channel_specific_paths(self):
+ self.points_directory = os.path.join(self.output_dir, "points")
+ self.detected_points = os.path.join(self.points_directory, "cells.xml")
+ self.classified_points = os.path.join(
+ self.points_directory, "cell_classification.xml"
+ )
+ self.downsampled_points = os.path.join(
+ self.points_directory, "downsampled.points"
+ )
+ self.atlas_points = os.path.join(self.points_directory, "atlas.points")
+ self.brainrender_points = os.path.join(
+ self.points_directory, "points.npy"
+ )
+ self.abc4d_points = os.path.join(self.points_directory, "abc4d.npy")
+
+ self.tmp__cubes_output_dir = os.path.join(self.output_dir, "cubes")
+
+ self.figures_directory = os.path.join(self.output_dir, "figures")
+ self.heatmap = os.path.join(self.figures_directory, "heatmap.tiff")
+
+ self.analysis_directory = os.path.join(self.output_dir, "analysis")
+ self.summary_csv = os.path.join(self.analysis_directory, "summary.csv")
+ self.all_points_csv = os.path.join(
+ self.analysis_directory, "all_points.csv"
+ )
+
+
+def serialise(obj):
+ if isinstance(obj, PurePath):
+ return str(obj)
+ else:
+ return obj.__dict__
+
+
+def log_metadata(file_path, args):
+ with open(file_path, "w") as f:
+ json.dump(args, f, default=serialise)
+
+
+def prep_cellfinder_general():
+ args = parser.cellfinder_parser().parse_args()
+ arg_groups = get_arg_groups(args, cellfinder_parser())
+
+ check_input_arg_existance(args)
+
+ if not os.path.exists(args.output_dir):
+ os.makedirs(args.output_dir)
+
+ args.paths = Paths(args.output_dir)
+
+ fancylog.start_logging(
+ args.output_dir,
+ program_for_log,
+ variables=[args, args.paths],
+ verbose=args.debug,
+ log_header="CELLFINDER LOG",
+ multiprocessing_aware=True,
+ )
+
+ log_metadata(args.paths.metadata_path, args)
+
+ what_to_run = CalcWhatToRun(args)
+ args.signal_ch_ids, args.background_ch_id = check_and_return_ch_ids(
+ args.signal_ch_ids, args.background_ch_id, args.signal_planes_paths
+ )
+ args.brainreg_paths = BrainRegPaths(args.paths.registration_output_folder)
+ atlas = BrainGlobeAtlas(args.atlas)
+ return args, arg_groups, what_to_run, atlas
+
+
+def check_and_return_ch_ids(signal_ids, background_id, signal_channel_list):
+ """
+ If channel IDs are provided (via CLI), then they are checked for
+ suitability (i.e. they are unique).
+ If channel IDs are not given, then unique IDs will be generated
+
+ Assumes only 1 background channel (for registration)
+
+ :param list signal_ids: List of ID ints, or None.
+ :param int background_id: ID int, or None.
+ :param list signal_channel_list: Input files
+ :return: list of signal channel IDs, and background channel ID
+ """
+
+ # TODO: make this more general, so it gives informative error messages
+ # when called from cellfinder.extract.cli
+
+ num_signal_channels = len(signal_channel_list)
+
+ if signal_ids is None:
+ if background_id is None:
+ signal_ids = list(range(num_signal_channels))
+ background_id = num_signal_channels # max(signal_ids) + 1
+ pass
+ else:
+ signal_start = background_id + 1
+ signal_end = signal_start + num_signal_channels
+ signal_ids = list(range(signal_start, signal_end))
+ pass
+ else:
+ check_correct_number_signal_channels(signal_ids, num_signal_channels)
+ check_signal_overlap(signal_ids)
+ if background_id is None:
+ background_id = max(signal_ids) + 1
+ pass
+ else:
+ # N.B. check_id_overlap needs bg_ch to be iterable (e.g. a list)
+ check_sig_bg_id_overlap(signal_ids, [background_id])
+ return signal_ids, background_id
+
+
+def check_correct_number_signal_channels(signal_ids, num_signal_channels):
+ num_signal_ids = len(signal_ids)
+ if num_signal_channels != num_signal_ids:
+ logging.error(
+ "Number of signal channel IDs ({}) does not match the number of "
+ "signal channels ({})".format(num_signal_ids, num_signal_channels)
+ )
+
+ raise CommandLineInputError(
+ "Number of signal channel IDs ({}) does not match the number of "
+ "signal channels ({})".format(num_signal_ids, num_signal_channels)
+ )
+
+
+def check_signal_overlap(signal_ids):
+ signal_unique, repeated_signal_ch = tools.check_unique_list(signal_ids)
+ if not signal_unique:
+ logging.error(
+ "Non-unique values for '--signal-channel-ids' ({}) given. ID:"
+ " {} given more than once.".format(signal_ids, repeated_signal_ch)
+ )
+ raise CommandLineInputError(
+ "Non-unique values for '--signal-channel-ids' ({}) given. ID:"
+ " {} given more than once.".format(signal_ids, repeated_signal_ch)
+ )
+
+
+def check_sig_bg_id_overlap(signal_ids, background_id):
+ common_id_result, intersection = tools.common_member(
+ signal_ids, background_id
+ )
+
+ if common_id_result:
+ logging.error(
+ "Non-unique values for '--signal-channel-ids' ({}) and for "
+ "'--background-channel-id' ({}) given. ID: {} was given in"
+ " both.".format(signal_ids, background_id, intersection)
+ )
+ raise CommandLineInputError(
+ "Non-unique values for '--signal-channel-ids' ({}) and for "
+ "'--background-channel-id' ({}) given. ID: {} was given in"
+ " both.".format(signal_ids, background_id, intersection)
+ )
+
+
+class CalcWhatToRun:
+ """
+ Class to (hopefully) simplify what should and shouldn't be run
+ """
+
+ def __init__(self, args):
+ self.detect = True
+ self.classify = True
+ self.register = True
+ self.analyse = True
+ self.figures = True
+ self.candidates_exist = True
+ self.cells_exist = True
+
+ self.atlas_image = os.path.join(
+ args.paths.registration_output_folder, "registered_atlas.tiff"
+ )
+ # order is important
+ self.cli_options(args)
+ self.existence()
+
+ def update(self, args):
+ self.cli_options(args)
+ self.existence()
+ self.channel_specific_update(args)
+
+ def cli_options(self, args):
+ self.detect = not args.no_detection
+ self.classify = not args.no_classification
+ self.register = not args.no_register
+ self.analyse = not args.no_analyse
+ self.figures = not args.no_figures
+
+ def existence(self):
+ if os.path.exists(self.atlas_image):
+ logging.warning(
+ "Registered atlas exists, assuming " "already run. Skipping."
+ )
+ self.register = False
+
+ def channel_specific_update(self, args):
+ if os.path.exists(args.paths.detected_points):
+ logging.warning(
+ "Initial detection file exists (cells.xml), "
+ "assuming already run. Skipping."
+ )
+ self.detect = False
+
+ if os.path.exists(args.paths.classified_points):
+ logging.warning(
+ "Cell classification file "
+ "(cell_classification.xml)"
+ " exists, assuming already run. Skipping cell detection, "
+ "and classification"
+ )
+ self.classify = False
+ self.detect = False
+
+ if not os.path.exists(self.atlas_image):
+ self.analyse = False
+ self.figures = False
+
+ if os.path.exists(args.paths.summary_csv):
+ self.analyse = False
+
+ if os.path.exists(args.paths.heatmap):
+ self.figures = False
+
+ def update_if_candidates_required(self):
+ if not self.candidates_exist:
+ self.classify = False
+
+ def update_if_cells_required(self):
+ if not self.cells_exist:
+ self.analyse = False
+ self.figures = False
+
+
+def prep_registration(args):
+ args.target_brain_path = args.background_planes_path[0]
+ logging.debug("Making registration directory")
+ ensure_directory_exists(args.paths.registration_output_folder)
+ log_metadata(args.paths.registration_metadata_path, args)
+ additional_images_downsample = {}
+ for idx, images in enumerate(args.signal_planes_paths):
+ channel = args.signal_ch_ids[idx]
+ additional_images_downsample[f"channel_{channel}"] = images
+ return args, additional_images_downsample
+
+
+def prep_channel_specific_general(args, what_to_run):
+ args.paths.output_dir = args.output_dir
+ args.paths.make_channel_specific_paths()
+ what_to_run.update(args)
+ return args, what_to_run
+
+
+def prep_candidate_detection(args):
+ if args.save_planes:
+ args.plane_directory = os.path.join(
+ args.output_dir, "processed_planes"
+ )
+ ensure_directory_exists(args.plane_directory)
+ else:
+ args.plane_directory = None # FIXME: remove this fudge
+
+ return args
diff --git a/cellfinder/tools/system.py b/cellfinder/tools/system.py
new file mode 100644
index 00000000..612ad4da
--- /dev/null
+++ b/cellfinder/tools/system.py
@@ -0,0 +1,82 @@
+from pathlib import Path
+
+from brainglobe_utils.general.exceptions import CommandLineInputError
+
+
+def get_subdirectories(directory, names_only=False):
+ """
+ Return all subdirectories in a given directory
+ :param directory:
+ :param names_only: If true, dont return paths, but the names
+ :return: Subdirectories
+ """
+
+ p = Path(directory)
+ if names_only:
+ return [x.name for x in p.iterdir() if x.is_dir()]
+ else:
+ return [x for x in p.iterdir() if x.is_dir()]
+
+
+def get_number_of_files_in_dir(directory):
+ """
+ Sums the number of files in a directory
+ :param directory: Any directory with files
+ :return: Number of files in directory
+ """
+ directory = Path(directory)
+ files = directory.iterdir()
+ total_files = sum(1 for x in files)
+ return total_files
+
+
+def check_path_exists(file):
+ """
+ Returns True is a file exists, otherwise throws a FileNotFoundError
+ :param file: Input file
+ :return: True, if the file exists
+ """
+ file = Path(file)
+ if file.exists():
+ return True
+ else:
+ raise FileNotFoundError
+
+
+def catch_input_file_error(path):
+ """
+ Catches if an input path doesn't exist, and returns an informative error
+ :param path: Input file path
+ default)
+ """
+ try:
+ check_path_exists(path)
+ except FileNotFoundError:
+ message = (
+ "File path: '{}' cannot be found. Please check your input "
+ "arguments.".format(path)
+ )
+ raise CommandLineInputError(message)
+
+
+def memory_in_bytes(memory_amount, unit):
+ """
+ Converts an amount of memory (in given units) to bytes
+
+ :param float memory_amount: An amount of memory
+ :param str unit: The units ('KB', 'MB', 'GB', 'TB', 'PB')
+ :return: Amount of memory in bytes
+ """
+ if memory_amount is None:
+ return memory_amount
+
+ supported_units = {"KB": 3, "MB": 6, "GB": 9, "TB": 12, "PB": 15}
+
+ unit = unit.upper()
+ if unit not in supported_units:
+ raise NotImplementedError(
+ f"Unit: {unit} is not supported. Please "
+ f"use one of {list(supported_units.keys())}"
+ )
+ else:
+ return memory_amount * 10 ** supported_units[unit]
diff --git a/cellfinder/tools/tools.py b/cellfinder/tools/tools.py
new file mode 100644
index 00000000..4d8431a5
--- /dev/null
+++ b/cellfinder/tools/tools.py
@@ -0,0 +1,126 @@
+from natsort import natsorted
+
+
+def get_max_value(obj_in):
+ """
+ Returns the maximum allowed value for a specific object type.
+ :param obj_in: Object
+ :return int: Maximum value of object type
+ """
+ # TODO: Generalise, and not rely on parsing a string
+ if obj_in.dtype == "uint8":
+ return 255
+ else:
+ return 2 ** int(str(obj_in.dtype)[-2:]) - 1
+
+
+def union(a, b):
+ """
+ Return the union (elements in both) of two lists
+ :param list a:
+ :param list b:
+ :return: Union of the two lists
+ """
+ return list(set(a) | set(b))
+
+
+def check_unique_list(in_list, natural_sort=True):
+ """
+ Checks if all the items in a list are unique or not
+ :param list in_list: Input list
+ :param bool natural_sort: Sort the resulting items naturally
+ (default: True)
+ :return: True/False and a list of any repeated values
+ """
+ unique = set(in_list)
+ repeated_items = []
+
+ for item in unique:
+ count = in_list.count(item)
+ if count > 1:
+ repeated_items.append(item)
+
+ if repeated_items:
+ if natural_sort:
+ repeated_items = natsorted(repeated_items)
+ return False, repeated_items
+ else:
+ return True, []
+
+
+def common_member(a, b, natural_sort=True):
+ """
+ Checks if two lists (or sets) have a common member, and if so, returns
+ the common members.
+ :param a: First list (or set)
+ :param b: Second list (or set)
+ :param bool natural_sort: Sort the resulting items naturally
+ (default: True)
+ :return: True/False and the list of values
+ """
+ a_set = set(a)
+ b_set = set(b)
+ intersection = list(a_set.intersection(b_set))
+ if len(intersection) > 0:
+ result = True
+ else:
+ result = False
+
+ if natural_sort:
+ intersection = natsorted(intersection)
+
+ return result, intersection
+
+
+def get_number_of_bins_nd(array_size, binning):
+ """
+ Generate the number of bins needed in three dimensions, based on the size
+ of the array, and the binning.
+ :param array_size: Size of the image array (tuple or dict)
+ :param binning: How many pixels in each edge of nD equal sized bins
+ :return:
+ """
+ if isinstance(array_size, tuple):
+ bins = [int(size / binning) for size in array_size]
+ elif isinstance(array_size, dict):
+ bins = [int(size / binning) for size in array_size.values()]
+ else:
+ raise NotImplementedError(
+ "Variable type: {} is not supported. Please"
+ "use tuple or dict".format(type(array_size))
+ )
+ return tuple(bins)
+
+
+def interchange_np_fiji_coordinates(tuple_in):
+ """
+ Swaps the first and second element of a tuple to swap between the numpy
+ convention (vertical is first) and FIJI convention (horizontal is first)
+ :param tuple_in: A 2+ element tuple
+ :return: Same tuple with element 0 and 1 interchanged
+ """
+ tmp_list = list(tuple_in)
+ tmp_list = swap_elements_list(tmp_list, 0, 1)
+ return tuple(tmp_list)
+
+
+def swap_elements_list(list_in, swap_a, swap_b):
+ """
+ Swap two elements in a list
+ :param list_in: A list
+ :param swap_a: Index of first element to swap
+ :param swap_b: Index of second element to swap
+ :return: List with swap_a and swap_b exchanged
+ """
+ list_in[swap_a], list_in[swap_b] = list_in[swap_b], list_in[swap_a]
+ return list_in
+
+
+def is_any_list_overlap(list_a, list_b):
+ """
+ Is there any overlap between two lists
+ :param list_a: Any list
+ :param list_b: Any other list
+ :return: True if lists have shared elements
+ """
+ return any({*list_a} & {*list_b})
diff --git a/environment.yml b/environment.yml
new file mode 100644
index 00000000..0691db0a
--- /dev/null
+++ b/environment.yml
@@ -0,0 +1,7 @@
+name: cellfinder
+dependencies:
+ - python=3.10
+ - cudatoolkit
+ - cudnn
+ - pip:
+ - cellfinder
diff --git a/pyproject.toml b/pyproject.toml
new file mode 100644
index 00000000..e589b61f
--- /dev/null
+++ b/pyproject.toml
@@ -0,0 +1,88 @@
+[project]
+name = "cellfinder"
+description = "Automated 3D cell detection and registration of whole-brain images"
+readme = "README.md"
+license = { file = "LICENSE" }
+requires-python = ">=3.8"
+authors = [
+ { name = "Adam Tyson", email = "code@adamltyson.com" },
+ { name = "Christian Niedworok" },
+ { name = "Charly Rousseau" },
+]
+classifiers = [
+ "Development Status :: 3 - Alpha",
+ "Operating System :: POSIX :: Linux",
+ "Operating System :: Microsoft :: Windows :: Windows 10",
+ "Programming Language :: Python",
+ "Programming Language :: Python :: 3.8",
+ "Programming Language :: Python :: 3.9",
+ "Programming Language :: Python :: 3.10",
+ "Topic :: Scientific/Engineering :: Image Recognition",
+ "Intended Audience :: Developers",
+ "Intended Audience :: Science/Research",
+]
+dependencies = [
+ "brainreg",
+ "cellfinder-core>=0.2.4",
+ "configobj",
+ "fancylog>=0.0.7",
+ "imio",
+ "brainglobe-utils>=0.2.5",
+ "multiprocessing-logging>=0.3.4",
+ "natsort",
+ "numpy",
+ "pandas",
+ "packaging",
+ "scikit-image",
+ "tifffile",
+ "tqdm",
+]
+dynamic = ["version"]
+
+[project.scripts]
+cellfinder = "cellfinder.main:main"
+
+[project.optional-dependencies]
+dev = [
+ "black",
+ "pytest-cov",
+ "pytest",
+ "gitpython",
+ "coverage>=5.0.3",
+ "pre-commit",
+ "setuptools_scm",
+]
+napari = ["napari[pyside2]", "brainglobe-napari-io", "cellfinder-napari"]
+
+[project.urls]
+source = "https://github.com/brainglobe/cellfinder"
+bug_tracker = "https://github.com/brainglobe/cellfinder/issues"
+homepage = "https://brainglobe.info"
+documentation = "https://brainglobe.info/documentation/cellfinder"
+[build-system]
+requires = ["setuptools>=45", "wheel", "setuptools_scm[toml]>=6.2"]
+build-backend = "setuptools.build_meta"
+
+[tool.black]
+target-version = ['py38', 'py39', 'py310']
+skip-string-normalization = false
+line-length = 79
+
+[tool.check-manifest]
+ignore = ["*.yaml"]
+
+[tool.ruff]
+line-length = 79
+exclude = ["__init__.py", "build", ".eggs"]
+select = ["I", "E", "F"]
+fix = true
+
+[tool.setuptools]
+include-package-data = true
+zip-safe = false
+
+[tool.setuptools.packages.find]
+include = ["cellfinder"]
+exclude = ["tests", "resources"]
+
+[tool.setuptools_scm]
diff --git a/pytest.ini b/pytest.ini
new file mode 100644
index 00000000..769170c1
--- /dev/null
+++ b/pytest.ini
@@ -0,0 +1,5 @@
+[pytest]
+addopts = --cov=cellfinder
+markers =
+ slow: marks tests as slow (deselect with '-m "not slow"')
+ serial
diff --git a/resources/brainrender.png b/resources/brainrender.png
new file mode 100644
index 00000000..66d510f5
Binary files /dev/null and b/resources/brainrender.png differ
diff --git a/resources/classify.png b/resources/classify.png
new file mode 100644
index 00000000..24c83454
Binary files /dev/null and b/resources/classify.png differ
diff --git a/resources/detect.png b/resources/detect.png
new file mode 100644
index 00000000..7ef4e5c9
Binary files /dev/null and b/resources/detect.png differ
diff --git a/resources/heatmap.png b/resources/heatmap.png
new file mode 100644
index 00000000..64df61af
Binary files /dev/null and b/resources/heatmap.png differ
diff --git a/resources/injection.png b/resources/injection.png
new file mode 100644
index 00000000..677fe1dd
Binary files /dev/null and b/resources/injection.png differ
diff --git a/resources/probe.png b/resources/probe.png
new file mode 100644
index 00000000..d894e909
Binary files /dev/null and b/resources/probe.png differ
diff --git a/resources/raw.png b/resources/raw.png
new file mode 100644
index 00000000..f2624947
Binary files /dev/null and b/resources/raw.png differ
diff --git a/resources/register.png b/resources/register.png
new file mode 100644
index 00000000..291fc69c
Binary files /dev/null and b/resources/register.png differ
diff --git a/tests/data/background/background_ch010000.tif b/tests/data/background/background_ch010000.tif
new file mode 100644
index 00000000..02e7313f
Binary files /dev/null and b/tests/data/background/background_ch010000.tif differ
diff --git a/tests/data/background/background_ch010001.tif b/tests/data/background/background_ch010001.tif
new file mode 100644
index 00000000..decf09bd
Binary files /dev/null and b/tests/data/background/background_ch010001.tif differ
diff --git a/tests/data/background/background_ch010002.tif b/tests/data/background/background_ch010002.tif
new file mode 100644
index 00000000..7c61cc72
Binary files /dev/null and b/tests/data/background/background_ch010002.tif differ
diff --git a/tests/data/background/background_ch010003.tif b/tests/data/background/background_ch010003.tif
new file mode 100644
index 00000000..5f501eaa
Binary files /dev/null and b/tests/data/background/background_ch010003.tif differ
diff --git a/tests/data/background/background_ch010004.tif b/tests/data/background/background_ch010004.tif
new file mode 100644
index 00000000..3871799e
Binary files /dev/null and b/tests/data/background/background_ch010004.tif differ
diff --git a/tests/data/background/background_ch010005.tif b/tests/data/background/background_ch010005.tif
new file mode 100644
index 00000000..6a502b95
Binary files /dev/null and b/tests/data/background/background_ch010005.tif differ
diff --git a/tests/data/background/background_ch010006.tif b/tests/data/background/background_ch010006.tif
new file mode 100644
index 00000000..09e6877b
Binary files /dev/null and b/tests/data/background/background_ch010006.tif differ
diff --git a/tests/data/background/background_ch010007.tif b/tests/data/background/background_ch010007.tif
new file mode 100644
index 00000000..ddce1149
Binary files /dev/null and b/tests/data/background/background_ch010007.tif differ
diff --git a/tests/data/background/background_ch010008.tif b/tests/data/background/background_ch010008.tif
new file mode 100644
index 00000000..46405368
Binary files /dev/null and b/tests/data/background/background_ch010008.tif differ
diff --git a/tests/data/background/background_ch010009.tif b/tests/data/background/background_ch010009.tif
new file mode 100644
index 00000000..c20619e7
Binary files /dev/null and b/tests/data/background/background_ch010009.tif differ
diff --git a/tests/data/background/background_ch010010.tif b/tests/data/background/background_ch010010.tif
new file mode 100644
index 00000000..daa78025
Binary files /dev/null and b/tests/data/background/background_ch010010.tif differ
diff --git a/tests/data/background/background_ch010011.tif b/tests/data/background/background_ch010011.tif
new file mode 100644
index 00000000..319342cf
Binary files /dev/null and b/tests/data/background/background_ch010011.tif differ
diff --git a/tests/data/background/background_ch010012.tif b/tests/data/background/background_ch010012.tif
new file mode 100644
index 00000000..26964f15
Binary files /dev/null and b/tests/data/background/background_ch010012.tif differ
diff --git a/tests/data/background/background_ch010013.tif b/tests/data/background/background_ch010013.tif
new file mode 100644
index 00000000..a1470754
Binary files /dev/null and b/tests/data/background/background_ch010013.tif differ
diff --git a/tests/data/background/background_ch010014.tif b/tests/data/background/background_ch010014.tif
new file mode 100644
index 00000000..873e57f1
Binary files /dev/null and b/tests/data/background/background_ch010014.tif differ
diff --git a/tests/data/background/background_ch010015.tif b/tests/data/background/background_ch010015.tif
new file mode 100644
index 00000000..8c48bade
Binary files /dev/null and b/tests/data/background/background_ch010015.tif differ
diff --git a/tests/data/background/background_ch010016.tif b/tests/data/background/background_ch010016.tif
new file mode 100644
index 00000000..afceb251
Binary files /dev/null and b/tests/data/background/background_ch010016.tif differ
diff --git a/tests/data/background/background_ch010017.tif b/tests/data/background/background_ch010017.tif
new file mode 100644
index 00000000..6c8f8f6e
Binary files /dev/null and b/tests/data/background/background_ch010017.tif differ
diff --git a/tests/data/background/background_ch010018.tif b/tests/data/background/background_ch010018.tif
new file mode 100644
index 00000000..71c6a578
Binary files /dev/null and b/tests/data/background/background_ch010018.tif differ
diff --git a/tests/data/background/background_ch010019.tif b/tests/data/background/background_ch010019.tif
new file mode 100644
index 00000000..17cd9891
Binary files /dev/null and b/tests/data/background/background_ch010019.tif differ
diff --git a/tests/data/background/background_ch010020.tif b/tests/data/background/background_ch010020.tif
new file mode 100644
index 00000000..db8f2999
Binary files /dev/null and b/tests/data/background/background_ch010020.tif differ
diff --git a/tests/data/background/background_ch010021.tif b/tests/data/background/background_ch010021.tif
new file mode 100644
index 00000000..c4cc1951
Binary files /dev/null and b/tests/data/background/background_ch010021.tif differ
diff --git a/tests/data/background/background_ch010022.tif b/tests/data/background/background_ch010022.tif
new file mode 100644
index 00000000..65cc4361
Binary files /dev/null and b/tests/data/background/background_ch010022.tif differ
diff --git a/tests/data/background/background_ch010023.tif b/tests/data/background/background_ch010023.tif
new file mode 100644
index 00000000..95c84061
Binary files /dev/null and b/tests/data/background/background_ch010023.tif differ
diff --git a/tests/data/background/background_ch010024.tif b/tests/data/background/background_ch010024.tif
new file mode 100644
index 00000000..6371ea2f
Binary files /dev/null and b/tests/data/background/background_ch010024.tif differ
diff --git a/tests/data/background/background_ch010025.tif b/tests/data/background/background_ch010025.tif
new file mode 100644
index 00000000..66e370ba
Binary files /dev/null and b/tests/data/background/background_ch010025.tif differ
diff --git a/tests/data/brain/image_0000.tif b/tests/data/brain/image_0000.tif
new file mode 100644
index 00000000..c1288515
Binary files /dev/null and b/tests/data/brain/image_0000.tif differ
diff --git a/tests/data/brain/image_0001.tif b/tests/data/brain/image_0001.tif
new file mode 100644
index 00000000..5c32402b
Binary files /dev/null and b/tests/data/brain/image_0001.tif differ
diff --git a/tests/data/brain/image_0002.tif b/tests/data/brain/image_0002.tif
new file mode 100644
index 00000000..cb9132bf
Binary files /dev/null and b/tests/data/brain/image_0002.tif differ
diff --git a/tests/data/brain/image_0003.tif b/tests/data/brain/image_0003.tif
new file mode 100644
index 00000000..573452a1
Binary files /dev/null and b/tests/data/brain/image_0003.tif differ
diff --git a/tests/data/brain/image_0004.tif b/tests/data/brain/image_0004.tif
new file mode 100644
index 00000000..c2f57e1b
Binary files /dev/null and b/tests/data/brain/image_0004.tif differ
diff --git a/tests/data/brain/image_0005.tif b/tests/data/brain/image_0005.tif
new file mode 100644
index 00000000..8c619b3d
Binary files /dev/null and b/tests/data/brain/image_0005.tif differ
diff --git a/tests/data/brain/image_0006.tif b/tests/data/brain/image_0006.tif
new file mode 100644
index 00000000..4457e3f0
Binary files /dev/null and b/tests/data/brain/image_0006.tif differ
diff --git a/tests/data/brain/image_0007.tif b/tests/data/brain/image_0007.tif
new file mode 100644
index 00000000..bf436e82
Binary files /dev/null and b/tests/data/brain/image_0007.tif differ
diff --git a/tests/data/brain/image_0008.tif b/tests/data/brain/image_0008.tif
new file mode 100644
index 00000000..e949c008
Binary files /dev/null and b/tests/data/brain/image_0008.tif differ
diff --git a/tests/data/brain/image_0009.tif b/tests/data/brain/image_0009.tif
new file mode 100644
index 00000000..f0e444a7
Binary files /dev/null and b/tests/data/brain/image_0009.tif differ
diff --git a/tests/data/brain/image_0010.tif b/tests/data/brain/image_0010.tif
new file mode 100644
index 00000000..39e4623c
Binary files /dev/null and b/tests/data/brain/image_0010.tif differ
diff --git a/tests/data/brain/image_0011.tif b/tests/data/brain/image_0011.tif
new file mode 100644
index 00000000..13487516
Binary files /dev/null and b/tests/data/brain/image_0011.tif differ
diff --git a/tests/data/brain/image_0012.tif b/tests/data/brain/image_0012.tif
new file mode 100644
index 00000000..1917add3
Binary files /dev/null and b/tests/data/brain/image_0012.tif differ
diff --git a/tests/data/brain/image_0013.tif b/tests/data/brain/image_0013.tif
new file mode 100644
index 00000000..1a976f3c
Binary files /dev/null and b/tests/data/brain/image_0013.tif differ
diff --git a/tests/data/brain/image_0014.tif b/tests/data/brain/image_0014.tif
new file mode 100644
index 00000000..eb469207
Binary files /dev/null and b/tests/data/brain/image_0014.tif differ
diff --git a/tests/data/brain/image_0015.tif b/tests/data/brain/image_0015.tif
new file mode 100644
index 00000000..804d4105
Binary files /dev/null and b/tests/data/brain/image_0015.tif differ
diff --git a/tests/data/brain/image_0016.tif b/tests/data/brain/image_0016.tif
new file mode 100644
index 00000000..c5ed95ac
Binary files /dev/null and b/tests/data/brain/image_0016.tif differ
diff --git a/tests/data/brain/image_0017.tif b/tests/data/brain/image_0017.tif
new file mode 100644
index 00000000..9bde06be
Binary files /dev/null and b/tests/data/brain/image_0017.tif differ
diff --git a/tests/data/brain/image_0018.tif b/tests/data/brain/image_0018.tif
new file mode 100644
index 00000000..be3b262d
Binary files /dev/null and b/tests/data/brain/image_0018.tif differ
diff --git a/tests/data/brain/image_0019.tif b/tests/data/brain/image_0019.tif
new file mode 100644
index 00000000..3542c9ba
Binary files /dev/null and b/tests/data/brain/image_0019.tif differ
diff --git a/tests/data/brain/image_0020.tif b/tests/data/brain/image_0020.tif
new file mode 100644
index 00000000..6eda4149
Binary files /dev/null and b/tests/data/brain/image_0020.tif differ
diff --git a/tests/data/brain/image_0021.tif b/tests/data/brain/image_0021.tif
new file mode 100644
index 00000000..a1fcddc1
Binary files /dev/null and b/tests/data/brain/image_0021.tif differ
diff --git a/tests/data/brain/image_0022.tif b/tests/data/brain/image_0022.tif
new file mode 100644
index 00000000..f1c4fe97
Binary files /dev/null and b/tests/data/brain/image_0022.tif differ
diff --git a/tests/data/brain/image_0023.tif b/tests/data/brain/image_0023.tif
new file mode 100644
index 00000000..3b5ed1c3
Binary files /dev/null and b/tests/data/brain/image_0023.tif differ
diff --git a/tests/data/brain/image_0024.tif b/tests/data/brain/image_0024.tif
new file mode 100644
index 00000000..11e3763c
Binary files /dev/null and b/tests/data/brain/image_0024.tif differ
diff --git a/tests/data/brain/image_0025.tif b/tests/data/brain/image_0025.tif
new file mode 100644
index 00000000..86d48deb
Binary files /dev/null and b/tests/data/brain/image_0025.tif differ
diff --git a/tests/data/brain/image_0026.tif b/tests/data/brain/image_0026.tif
new file mode 100644
index 00000000..90619430
Binary files /dev/null and b/tests/data/brain/image_0026.tif differ
diff --git a/tests/data/brain/image_0027.tif b/tests/data/brain/image_0027.tif
new file mode 100644
index 00000000..d3e0a784
Binary files /dev/null and b/tests/data/brain/image_0027.tif differ
diff --git a/tests/data/brain/image_0028.tif b/tests/data/brain/image_0028.tif
new file mode 100644
index 00000000..b84f7273
Binary files /dev/null and b/tests/data/brain/image_0028.tif differ
diff --git a/tests/data/brain/image_0029.tif b/tests/data/brain/image_0029.tif
new file mode 100644
index 00000000..c1fe51f2
Binary files /dev/null and b/tests/data/brain/image_0029.tif differ
diff --git a/tests/data/brain/image_0030.tif b/tests/data/brain/image_0030.tif
new file mode 100644
index 00000000..6723656b
Binary files /dev/null and b/tests/data/brain/image_0030.tif differ
diff --git a/tests/data/brain/image_0031.tif b/tests/data/brain/image_0031.tif
new file mode 100644
index 00000000..37d02908
Binary files /dev/null and b/tests/data/brain/image_0031.tif differ
diff --git a/tests/data/brain/image_0032.tif b/tests/data/brain/image_0032.tif
new file mode 100644
index 00000000..5e5fb256
Binary files /dev/null and b/tests/data/brain/image_0032.tif differ
diff --git a/tests/data/brain/image_0033.tif b/tests/data/brain/image_0033.tif
new file mode 100644
index 00000000..c9bb16ba
Binary files /dev/null and b/tests/data/brain/image_0033.tif differ
diff --git a/tests/data/brain/image_0034.tif b/tests/data/brain/image_0034.tif
new file mode 100644
index 00000000..0d24e31e
Binary files /dev/null and b/tests/data/brain/image_0034.tif differ
diff --git a/tests/data/brain/image_0035.tif b/tests/data/brain/image_0035.tif
new file mode 100644
index 00000000..c2062176
Binary files /dev/null and b/tests/data/brain/image_0035.tif differ
diff --git a/tests/data/brain/image_0036.tif b/tests/data/brain/image_0036.tif
new file mode 100644
index 00000000..d3d06625
Binary files /dev/null and b/tests/data/brain/image_0036.tif differ
diff --git a/tests/data/brain/image_0037.tif b/tests/data/brain/image_0037.tif
new file mode 100644
index 00000000..729cfd60
Binary files /dev/null and b/tests/data/brain/image_0037.tif differ
diff --git a/tests/data/brain/image_0038.tif b/tests/data/brain/image_0038.tif
new file mode 100644
index 00000000..1499f409
Binary files /dev/null and b/tests/data/brain/image_0038.tif differ
diff --git a/tests/data/brain/image_0039.tif b/tests/data/brain/image_0039.tif
new file mode 100644
index 00000000..67c32688
Binary files /dev/null and b/tests/data/brain/image_0039.tif differ
diff --git a/tests/data/brain/image_0040.tif b/tests/data/brain/image_0040.tif
new file mode 100644
index 00000000..7e3e89dd
Binary files /dev/null and b/tests/data/brain/image_0040.tif differ
diff --git a/tests/data/brain/image_0041.tif b/tests/data/brain/image_0041.tif
new file mode 100644
index 00000000..469cd6af
Binary files /dev/null and b/tests/data/brain/image_0041.tif differ
diff --git a/tests/data/brain/image_0042.tif b/tests/data/brain/image_0042.tif
new file mode 100644
index 00000000..9cd5ebe7
Binary files /dev/null and b/tests/data/brain/image_0042.tif differ
diff --git a/tests/data/brain/image_0043.tif b/tests/data/brain/image_0043.tif
new file mode 100644
index 00000000..a23adaf6
Binary files /dev/null and b/tests/data/brain/image_0043.tif differ
diff --git a/tests/data/brain/image_0044.tif b/tests/data/brain/image_0044.tif
new file mode 100644
index 00000000..8ee8bbee
Binary files /dev/null and b/tests/data/brain/image_0044.tif differ
diff --git a/tests/data/brain/image_0045.tif b/tests/data/brain/image_0045.tif
new file mode 100644
index 00000000..be724a3b
Binary files /dev/null and b/tests/data/brain/image_0045.tif differ
diff --git a/tests/data/brain/image_0046.tif b/tests/data/brain/image_0046.tif
new file mode 100644
index 00000000..b5863cbe
Binary files /dev/null and b/tests/data/brain/image_0046.tif differ
diff --git a/tests/data/brain/image_0047.tif b/tests/data/brain/image_0047.tif
new file mode 100644
index 00000000..2cf23f5e
Binary files /dev/null and b/tests/data/brain/image_0047.tif differ
diff --git a/tests/data/brain/image_0048.tif b/tests/data/brain/image_0048.tif
new file mode 100644
index 00000000..a7e1db4a
Binary files /dev/null and b/tests/data/brain/image_0048.tif differ
diff --git a/tests/data/brain/image_0049.tif b/tests/data/brain/image_0049.tif
new file mode 100644
index 00000000..e19142ee
Binary files /dev/null and b/tests/data/brain/image_0049.tif differ
diff --git a/tests/data/brain/image_0050.tif b/tests/data/brain/image_0050.tif
new file mode 100644
index 00000000..7f4ed5d0
Binary files /dev/null and b/tests/data/brain/image_0050.tif differ
diff --git a/tests/data/brain/image_0051.tif b/tests/data/brain/image_0051.tif
new file mode 100644
index 00000000..cfeacfb5
Binary files /dev/null and b/tests/data/brain/image_0051.tif differ
diff --git a/tests/data/brain/image_0052.tif b/tests/data/brain/image_0052.tif
new file mode 100644
index 00000000..71aaf4d1
Binary files /dev/null and b/tests/data/brain/image_0052.tif differ
diff --git a/tests/data/brain/image_0053.tif b/tests/data/brain/image_0053.tif
new file mode 100644
index 00000000..f5aa6218
Binary files /dev/null and b/tests/data/brain/image_0053.tif differ
diff --git a/tests/data/brain/image_0054.tif b/tests/data/brain/image_0054.tif
new file mode 100644
index 00000000..b7e82d95
Binary files /dev/null and b/tests/data/brain/image_0054.tif differ
diff --git a/tests/data/brain/image_0055.tif b/tests/data/brain/image_0055.tif
new file mode 100644
index 00000000..2c30c083
Binary files /dev/null and b/tests/data/brain/image_0055.tif differ
diff --git a/tests/data/brain/image_0056.tif b/tests/data/brain/image_0056.tif
new file mode 100644
index 00000000..b8e69938
Binary files /dev/null and b/tests/data/brain/image_0056.tif differ
diff --git a/tests/data/brain/image_0057.tif b/tests/data/brain/image_0057.tif
new file mode 100644
index 00000000..489d14db
Binary files /dev/null and b/tests/data/brain/image_0057.tif differ
diff --git a/tests/data/brain/image_0058.tif b/tests/data/brain/image_0058.tif
new file mode 100644
index 00000000..258da06d
Binary files /dev/null and b/tests/data/brain/image_0058.tif differ
diff --git a/tests/data/brain/image_0059.tif b/tests/data/brain/image_0059.tif
new file mode 100644
index 00000000..e38be562
Binary files /dev/null and b/tests/data/brain/image_0059.tif differ
diff --git a/tests/data/brain/image_0060.tif b/tests/data/brain/image_0060.tif
new file mode 100644
index 00000000..629f057c
Binary files /dev/null and b/tests/data/brain/image_0060.tif differ
diff --git a/tests/data/brain/image_0061.tif b/tests/data/brain/image_0061.tif
new file mode 100644
index 00000000..bc5d969e
Binary files /dev/null and b/tests/data/brain/image_0061.tif differ
diff --git a/tests/data/brain/image_0062.tif b/tests/data/brain/image_0062.tif
new file mode 100644
index 00000000..8dd288bd
Binary files /dev/null and b/tests/data/brain/image_0062.tif differ
diff --git a/tests/data/brain/image_0063.tif b/tests/data/brain/image_0063.tif
new file mode 100644
index 00000000..1db3e696
Binary files /dev/null and b/tests/data/brain/image_0063.tif differ
diff --git a/tests/data/brain/image_0064.tif b/tests/data/brain/image_0064.tif
new file mode 100644
index 00000000..0b620f47
Binary files /dev/null and b/tests/data/brain/image_0064.tif differ
diff --git a/tests/data/brain/image_0065.tif b/tests/data/brain/image_0065.tif
new file mode 100644
index 00000000..e88e829b
Binary files /dev/null and b/tests/data/brain/image_0065.tif differ
diff --git a/tests/data/brain/image_0066.tif b/tests/data/brain/image_0066.tif
new file mode 100644
index 00000000..a5e5f1ab
Binary files /dev/null and b/tests/data/brain/image_0066.tif differ
diff --git a/tests/data/brain/image_0067.tif b/tests/data/brain/image_0067.tif
new file mode 100644
index 00000000..88159e37
Binary files /dev/null and b/tests/data/brain/image_0067.tif differ
diff --git a/tests/data/brain/image_0068.tif b/tests/data/brain/image_0068.tif
new file mode 100644
index 00000000..ae6a0ccc
Binary files /dev/null and b/tests/data/brain/image_0068.tif differ
diff --git a/tests/data/brain/image_0069.tif b/tests/data/brain/image_0069.tif
new file mode 100644
index 00000000..6124f409
Binary files /dev/null and b/tests/data/brain/image_0069.tif differ
diff --git a/tests/data/brain/image_0070.tif b/tests/data/brain/image_0070.tif
new file mode 100644
index 00000000..fea1c7ab
Binary files /dev/null and b/tests/data/brain/image_0070.tif differ
diff --git a/tests/data/brain/image_0071.tif b/tests/data/brain/image_0071.tif
new file mode 100644
index 00000000..bc643608
Binary files /dev/null and b/tests/data/brain/image_0071.tif differ
diff --git a/tests/data/brain/image_0072.tif b/tests/data/brain/image_0072.tif
new file mode 100644
index 00000000..0b23a9b0
Binary files /dev/null and b/tests/data/brain/image_0072.tif differ
diff --git a/tests/data/brain/image_0073.tif b/tests/data/brain/image_0073.tif
new file mode 100644
index 00000000..10290e41
Binary files /dev/null and b/tests/data/brain/image_0073.tif differ
diff --git a/tests/data/brain/image_0074.tif b/tests/data/brain/image_0074.tif
new file mode 100644
index 00000000..dd990b13
Binary files /dev/null and b/tests/data/brain/image_0074.tif differ
diff --git a/tests/data/brain/image_0075.tif b/tests/data/brain/image_0075.tif
new file mode 100644
index 00000000..2c4bd305
Binary files /dev/null and b/tests/data/brain/image_0075.tif differ
diff --git a/tests/data/brain/image_0076.tif b/tests/data/brain/image_0076.tif
new file mode 100644
index 00000000..6a3b3c60
Binary files /dev/null and b/tests/data/brain/image_0076.tif differ
diff --git a/tests/data/brain/image_0077.tif b/tests/data/brain/image_0077.tif
new file mode 100644
index 00000000..8a979db1
Binary files /dev/null and b/tests/data/brain/image_0077.tif differ
diff --git a/tests/data/brain/image_0078.tif b/tests/data/brain/image_0078.tif
new file mode 100644
index 00000000..61d4b34c
Binary files /dev/null and b/tests/data/brain/image_0078.tif differ
diff --git a/tests/data/brain/image_0079.tif b/tests/data/brain/image_0079.tif
new file mode 100644
index 00000000..78634a33
Binary files /dev/null and b/tests/data/brain/image_0079.tif differ
diff --git a/tests/data/brain/image_0080.tif b/tests/data/brain/image_0080.tif
new file mode 100644
index 00000000..7916e02a
Binary files /dev/null and b/tests/data/brain/image_0080.tif differ
diff --git a/tests/data/brain/image_0081.tif b/tests/data/brain/image_0081.tif
new file mode 100644
index 00000000..79019bbd
Binary files /dev/null and b/tests/data/brain/image_0081.tif differ
diff --git a/tests/data/brain/image_0082.tif b/tests/data/brain/image_0082.tif
new file mode 100644
index 00000000..075fc1c6
Binary files /dev/null and b/tests/data/brain/image_0082.tif differ
diff --git a/tests/data/brain/image_0083.tif b/tests/data/brain/image_0083.tif
new file mode 100644
index 00000000..d2a38579
Binary files /dev/null and b/tests/data/brain/image_0083.tif differ
diff --git a/tests/data/brain/image_0084.tif b/tests/data/brain/image_0084.tif
new file mode 100644
index 00000000..eef137c9
Binary files /dev/null and b/tests/data/brain/image_0084.tif differ
diff --git a/tests/data/brain/image_0085.tif b/tests/data/brain/image_0085.tif
new file mode 100644
index 00000000..6524db0f
Binary files /dev/null and b/tests/data/brain/image_0085.tif differ
diff --git a/tests/data/brain/image_0086.tif b/tests/data/brain/image_0086.tif
new file mode 100644
index 00000000..6cd99e60
Binary files /dev/null and b/tests/data/brain/image_0086.tif differ
diff --git a/tests/data/brain/image_0087.tif b/tests/data/brain/image_0087.tif
new file mode 100644
index 00000000..54f6c7f6
Binary files /dev/null and b/tests/data/brain/image_0087.tif differ
diff --git a/tests/data/brain/image_0088.tif b/tests/data/brain/image_0088.tif
new file mode 100644
index 00000000..cefe85eb
Binary files /dev/null and b/tests/data/brain/image_0088.tif differ
diff --git a/tests/data/brain/image_0089.tif b/tests/data/brain/image_0089.tif
new file mode 100644
index 00000000..6a6c4480
Binary files /dev/null and b/tests/data/brain/image_0089.tif differ
diff --git a/tests/data/brain/image_0090.tif b/tests/data/brain/image_0090.tif
new file mode 100644
index 00000000..4f74cfa8
Binary files /dev/null and b/tests/data/brain/image_0090.tif differ
diff --git a/tests/data/brain/image_0091.tif b/tests/data/brain/image_0091.tif
new file mode 100644
index 00000000..d64f19ab
Binary files /dev/null and b/tests/data/brain/image_0091.tif differ
diff --git a/tests/data/brain/image_0092.tif b/tests/data/brain/image_0092.tif
new file mode 100644
index 00000000..85c86525
Binary files /dev/null and b/tests/data/brain/image_0092.tif differ
diff --git a/tests/data/brain/image_0093.tif b/tests/data/brain/image_0093.tif
new file mode 100644
index 00000000..874748e1
Binary files /dev/null and b/tests/data/brain/image_0093.tif differ
diff --git a/tests/data/brain/image_0094.tif b/tests/data/brain/image_0094.tif
new file mode 100644
index 00000000..c07840ae
Binary files /dev/null and b/tests/data/brain/image_0094.tif differ
diff --git a/tests/data/brain/image_0095.tif b/tests/data/brain/image_0095.tif
new file mode 100644
index 00000000..c6c85faa
Binary files /dev/null and b/tests/data/brain/image_0095.tif differ
diff --git a/tests/data/brain/image_0096.tif b/tests/data/brain/image_0096.tif
new file mode 100644
index 00000000..5b1f7d3e
Binary files /dev/null and b/tests/data/brain/image_0096.tif differ
diff --git a/tests/data/brain/image_0097.tif b/tests/data/brain/image_0097.tif
new file mode 100644
index 00000000..38e3440a
Binary files /dev/null and b/tests/data/brain/image_0097.tif differ
diff --git a/tests/data/brain/image_0098.tif b/tests/data/brain/image_0098.tif
new file mode 100644
index 00000000..89302f5d
Binary files /dev/null and b/tests/data/brain/image_0098.tif differ
diff --git a/tests/data/brain/image_0099.tif b/tests/data/brain/image_0099.tif
new file mode 100644
index 00000000..6538147c
Binary files /dev/null and b/tests/data/brain/image_0099.tif differ
diff --git a/tests/data/brain/image_0100.tif b/tests/data/brain/image_0100.tif
new file mode 100644
index 00000000..9d7a29a1
Binary files /dev/null and b/tests/data/brain/image_0100.tif differ
diff --git a/tests/data/brain/image_0101.tif b/tests/data/brain/image_0101.tif
new file mode 100644
index 00000000..6e2dde74
Binary files /dev/null and b/tests/data/brain/image_0101.tif differ
diff --git a/tests/data/brain/image_0102.tif b/tests/data/brain/image_0102.tif
new file mode 100644
index 00000000..cb2e79f9
Binary files /dev/null and b/tests/data/brain/image_0102.tif differ
diff --git a/tests/data/brain/image_0103.tif b/tests/data/brain/image_0103.tif
new file mode 100644
index 00000000..63b33730
Binary files /dev/null and b/tests/data/brain/image_0103.tif differ
diff --git a/tests/data/brain/image_0104.tif b/tests/data/brain/image_0104.tif
new file mode 100644
index 00000000..c542e061
Binary files /dev/null and b/tests/data/brain/image_0104.tif differ
diff --git a/tests/data/brain/image_0105.tif b/tests/data/brain/image_0105.tif
new file mode 100644
index 00000000..19a43ac1
Binary files /dev/null and b/tests/data/brain/image_0105.tif differ
diff --git a/tests/data/brain/image_0106.tif b/tests/data/brain/image_0106.tif
new file mode 100644
index 00000000..f8bd91ae
Binary files /dev/null and b/tests/data/brain/image_0106.tif differ
diff --git a/tests/data/brain/image_0107.tif b/tests/data/brain/image_0107.tif
new file mode 100644
index 00000000..955fa6be
Binary files /dev/null and b/tests/data/brain/image_0107.tif differ
diff --git a/tests/data/brain/image_0108.tif b/tests/data/brain/image_0108.tif
new file mode 100644
index 00000000..f5a01fef
Binary files /dev/null and b/tests/data/brain/image_0108.tif differ
diff --git a/tests/data/brain/image_0109.tif b/tests/data/brain/image_0109.tif
new file mode 100644
index 00000000..2a0cd4fc
Binary files /dev/null and b/tests/data/brain/image_0109.tif differ
diff --git a/tests/data/brain/image_0110.tif b/tests/data/brain/image_0110.tif
new file mode 100644
index 00000000..4544ba10
Binary files /dev/null and b/tests/data/brain/image_0110.tif differ
diff --git a/tests/data/brain/image_0111.tif b/tests/data/brain/image_0111.tif
new file mode 100644
index 00000000..d4fb8650
Binary files /dev/null and b/tests/data/brain/image_0111.tif differ
diff --git a/tests/data/brain/image_0112.tif b/tests/data/brain/image_0112.tif
new file mode 100644
index 00000000..98a56d77
Binary files /dev/null and b/tests/data/brain/image_0112.tif differ
diff --git a/tests/data/brain/image_0113.tif b/tests/data/brain/image_0113.tif
new file mode 100644
index 00000000..781561f8
Binary files /dev/null and b/tests/data/brain/image_0113.tif differ
diff --git a/tests/data/brain/image_0114.tif b/tests/data/brain/image_0114.tif
new file mode 100644
index 00000000..e48dd0b5
Binary files /dev/null and b/tests/data/brain/image_0114.tif differ
diff --git a/tests/data/brain/image_0115.tif b/tests/data/brain/image_0115.tif
new file mode 100644
index 00000000..5b799bab
Binary files /dev/null and b/tests/data/brain/image_0115.tif differ
diff --git a/tests/data/brain/image_0116.tif b/tests/data/brain/image_0116.tif
new file mode 100644
index 00000000..25ac832d
Binary files /dev/null and b/tests/data/brain/image_0116.tif differ
diff --git a/tests/data/brain/image_0117.tif b/tests/data/brain/image_0117.tif
new file mode 100644
index 00000000..fdba09b9
Binary files /dev/null and b/tests/data/brain/image_0117.tif differ
diff --git a/tests/data/brain/image_0118.tif b/tests/data/brain/image_0118.tif
new file mode 100644
index 00000000..5e94f59c
Binary files /dev/null and b/tests/data/brain/image_0118.tif differ
diff --git a/tests/data/brain/image_0119.tif b/tests/data/brain/image_0119.tif
new file mode 100644
index 00000000..7398b364
Binary files /dev/null and b/tests/data/brain/image_0119.tif differ
diff --git a/tests/data/brain/image_0120.tif b/tests/data/brain/image_0120.tif
new file mode 100644
index 00000000..94e923d0
Binary files /dev/null and b/tests/data/brain/image_0120.tif differ
diff --git a/tests/data/brain/image_0121.tif b/tests/data/brain/image_0121.tif
new file mode 100644
index 00000000..be3fd64a
Binary files /dev/null and b/tests/data/brain/image_0121.tif differ
diff --git a/tests/data/brain/image_0122.tif b/tests/data/brain/image_0122.tif
new file mode 100644
index 00000000..d98260c7
Binary files /dev/null and b/tests/data/brain/image_0122.tif differ
diff --git a/tests/data/brain/image_0123.tif b/tests/data/brain/image_0123.tif
new file mode 100644
index 00000000..5d88e5bf
Binary files /dev/null and b/tests/data/brain/image_0123.tif differ
diff --git a/tests/data/brain/image_0124.tif b/tests/data/brain/image_0124.tif
new file mode 100644
index 00000000..9e460d51
Binary files /dev/null and b/tests/data/brain/image_0124.tif differ
diff --git a/tests/data/brain/image_0125.tif b/tests/data/brain/image_0125.tif
new file mode 100644
index 00000000..ca4dd1c8
Binary files /dev/null and b/tests/data/brain/image_0125.tif differ
diff --git a/tests/data/brain/image_0126.tif b/tests/data/brain/image_0126.tif
new file mode 100644
index 00000000..cde39d1a
Binary files /dev/null and b/tests/data/brain/image_0126.tif differ
diff --git a/tests/data/brain/image_0127.tif b/tests/data/brain/image_0127.tif
new file mode 100644
index 00000000..33b19c75
Binary files /dev/null and b/tests/data/brain/image_0127.tif differ
diff --git a/tests/data/brain/image_0128.tif b/tests/data/brain/image_0128.tif
new file mode 100644
index 00000000..8c791fa4
Binary files /dev/null and b/tests/data/brain/image_0128.tif differ
diff --git a/tests/data/brain/image_0129.tif b/tests/data/brain/image_0129.tif
new file mode 100644
index 00000000..e14bb12e
Binary files /dev/null and b/tests/data/brain/image_0129.tif differ
diff --git a/tests/data/brain/image_0130.tif b/tests/data/brain/image_0130.tif
new file mode 100644
index 00000000..a4ba96e7
Binary files /dev/null and b/tests/data/brain/image_0130.tif differ
diff --git a/tests/data/brain/image_0131.tif b/tests/data/brain/image_0131.tif
new file mode 100644
index 00000000..69ff721d
Binary files /dev/null and b/tests/data/brain/image_0131.tif differ
diff --git a/tests/data/brain/image_0132.tif b/tests/data/brain/image_0132.tif
new file mode 100644
index 00000000..e922be5f
Binary files /dev/null and b/tests/data/brain/image_0132.tif differ
diff --git a/tests/data/brain/image_0133.tif b/tests/data/brain/image_0133.tif
new file mode 100644
index 00000000..e17bf60e
Binary files /dev/null and b/tests/data/brain/image_0133.tif differ
diff --git a/tests/data/brain/image_0134.tif b/tests/data/brain/image_0134.tif
new file mode 100644
index 00000000..e7937bbe
Binary files /dev/null and b/tests/data/brain/image_0134.tif differ
diff --git a/tests/data/brain/image_0135.tif b/tests/data/brain/image_0135.tif
new file mode 100644
index 00000000..28f80157
Binary files /dev/null and b/tests/data/brain/image_0135.tif differ
diff --git a/tests/data/brain/image_0136.tif b/tests/data/brain/image_0136.tif
new file mode 100644
index 00000000..000581b2
Binary files /dev/null and b/tests/data/brain/image_0136.tif differ
diff --git a/tests/data/brain/image_0137.tif b/tests/data/brain/image_0137.tif
new file mode 100644
index 00000000..70fae025
Binary files /dev/null and b/tests/data/brain/image_0137.tif differ
diff --git a/tests/data/brain/image_0138.tif b/tests/data/brain/image_0138.tif
new file mode 100644
index 00000000..b2e2b5ea
Binary files /dev/null and b/tests/data/brain/image_0138.tif differ
diff --git a/tests/data/brain/image_0139.tif b/tests/data/brain/image_0139.tif
new file mode 100644
index 00000000..ffcfd2d6
Binary files /dev/null and b/tests/data/brain/image_0139.tif differ
diff --git a/tests/data/brain/image_0140.tif b/tests/data/brain/image_0140.tif
new file mode 100644
index 00000000..1498893e
Binary files /dev/null and b/tests/data/brain/image_0140.tif differ
diff --git a/tests/data/brain/image_0141.tif b/tests/data/brain/image_0141.tif
new file mode 100644
index 00000000..98ef5f8b
Binary files /dev/null and b/tests/data/brain/image_0141.tif differ
diff --git a/tests/data/brain/image_0142.tif b/tests/data/brain/image_0142.tif
new file mode 100644
index 00000000..bdedd289
Binary files /dev/null and b/tests/data/brain/image_0142.tif differ
diff --git a/tests/data/brain/image_0143.tif b/tests/data/brain/image_0143.tif
new file mode 100644
index 00000000..5c3d23a5
Binary files /dev/null and b/tests/data/brain/image_0143.tif differ
diff --git a/tests/data/brain/image_0144.tif b/tests/data/brain/image_0144.tif
new file mode 100644
index 00000000..06025c06
Binary files /dev/null and b/tests/data/brain/image_0144.tif differ
diff --git a/tests/data/brain/image_0145.tif b/tests/data/brain/image_0145.tif
new file mode 100644
index 00000000..13007f2d
Binary files /dev/null and b/tests/data/brain/image_0145.tif differ
diff --git a/tests/data/brain/image_0146.tif b/tests/data/brain/image_0146.tif
new file mode 100644
index 00000000..a92c7aa7
Binary files /dev/null and b/tests/data/brain/image_0146.tif differ
diff --git a/tests/data/brain/image_0147.tif b/tests/data/brain/image_0147.tif
new file mode 100644
index 00000000..7a10becf
Binary files /dev/null and b/tests/data/brain/image_0147.tif differ
diff --git a/tests/data/brain/image_0148.tif b/tests/data/brain/image_0148.tif
new file mode 100644
index 00000000..6830a952
Binary files /dev/null and b/tests/data/brain/image_0148.tif differ
diff --git a/tests/data/brain/image_0149.tif b/tests/data/brain/image_0149.tif
new file mode 100644
index 00000000..34064045
Binary files /dev/null and b/tests/data/brain/image_0149.tif differ
diff --git a/tests/data/brain/image_0150.tif b/tests/data/brain/image_0150.tif
new file mode 100644
index 00000000..09ac0b9b
Binary files /dev/null and b/tests/data/brain/image_0150.tif differ
diff --git a/tests/data/brain/image_0151.tif b/tests/data/brain/image_0151.tif
new file mode 100644
index 00000000..62c1f916
Binary files /dev/null and b/tests/data/brain/image_0151.tif differ
diff --git a/tests/data/brain/image_0152.tif b/tests/data/brain/image_0152.tif
new file mode 100644
index 00000000..6b084d4f
Binary files /dev/null and b/tests/data/brain/image_0152.tif differ
diff --git a/tests/data/brain/image_0153.tif b/tests/data/brain/image_0153.tif
new file mode 100644
index 00000000..d6a72348
Binary files /dev/null and b/tests/data/brain/image_0153.tif differ
diff --git a/tests/data/brain/image_0154.tif b/tests/data/brain/image_0154.tif
new file mode 100644
index 00000000..6b113878
Binary files /dev/null and b/tests/data/brain/image_0154.tif differ
diff --git a/tests/data/brain/image_0155.tif b/tests/data/brain/image_0155.tif
new file mode 100644
index 00000000..9f192a8d
Binary files /dev/null and b/tests/data/brain/image_0155.tif differ
diff --git a/tests/data/brain/image_0156.tif b/tests/data/brain/image_0156.tif
new file mode 100644
index 00000000..3b9d998f
Binary files /dev/null and b/tests/data/brain/image_0156.tif differ
diff --git a/tests/data/brain/image_0157.tif b/tests/data/brain/image_0157.tif
new file mode 100644
index 00000000..872ddf8d
Binary files /dev/null and b/tests/data/brain/image_0157.tif differ
diff --git a/tests/data/brain/image_0158.tif b/tests/data/brain/image_0158.tif
new file mode 100644
index 00000000..5f5302d6
Binary files /dev/null and b/tests/data/brain/image_0158.tif differ
diff --git a/tests/data/brain/image_0159.tif b/tests/data/brain/image_0159.tif
new file mode 100644
index 00000000..5d9cf185
Binary files /dev/null and b/tests/data/brain/image_0159.tif differ
diff --git a/tests/data/brain/image_0160.tif b/tests/data/brain/image_0160.tif
new file mode 100644
index 00000000..4769fd91
Binary files /dev/null and b/tests/data/brain/image_0160.tif differ
diff --git a/tests/data/brain/image_0161.tif b/tests/data/brain/image_0161.tif
new file mode 100644
index 00000000..5847adf6
Binary files /dev/null and b/tests/data/brain/image_0161.tif differ
diff --git a/tests/data/brain/image_0162.tif b/tests/data/brain/image_0162.tif
new file mode 100644
index 00000000..8503e192
Binary files /dev/null and b/tests/data/brain/image_0162.tif differ
diff --git a/tests/data/brain/image_0163.tif b/tests/data/brain/image_0163.tif
new file mode 100644
index 00000000..c3d8e561
Binary files /dev/null and b/tests/data/brain/image_0163.tif differ
diff --git a/tests/data/brain/image_0164.tif b/tests/data/brain/image_0164.tif
new file mode 100644
index 00000000..5c0486ea
Binary files /dev/null and b/tests/data/brain/image_0164.tif differ
diff --git a/tests/data/brain/image_0165.tif b/tests/data/brain/image_0165.tif
new file mode 100644
index 00000000..b8e3488a
Binary files /dev/null and b/tests/data/brain/image_0165.tif differ
diff --git a/tests/data/brain/image_0166.tif b/tests/data/brain/image_0166.tif
new file mode 100644
index 00000000..c82475e6
Binary files /dev/null and b/tests/data/brain/image_0166.tif differ
diff --git a/tests/data/brain/image_0167.tif b/tests/data/brain/image_0167.tif
new file mode 100644
index 00000000..4eeb5ad4
Binary files /dev/null and b/tests/data/brain/image_0167.tif differ
diff --git a/tests/data/brain/image_0168.tif b/tests/data/brain/image_0168.tif
new file mode 100644
index 00000000..d9bb6d9b
Binary files /dev/null and b/tests/data/brain/image_0168.tif differ
diff --git a/tests/data/brain/image_0169.tif b/tests/data/brain/image_0169.tif
new file mode 100644
index 00000000..b1f81791
Binary files /dev/null and b/tests/data/brain/image_0169.tif differ
diff --git a/tests/data/brain/image_0170.tif b/tests/data/brain/image_0170.tif
new file mode 100644
index 00000000..f968a623
Binary files /dev/null and b/tests/data/brain/image_0170.tif differ
diff --git a/tests/data/brain/image_0171.tif b/tests/data/brain/image_0171.tif
new file mode 100644
index 00000000..f84f7b8c
Binary files /dev/null and b/tests/data/brain/image_0171.tif differ
diff --git a/tests/data/brain/image_0172.tif b/tests/data/brain/image_0172.tif
new file mode 100644
index 00000000..a581d39d
Binary files /dev/null and b/tests/data/brain/image_0172.tif differ
diff --git a/tests/data/brain/image_0173.tif b/tests/data/brain/image_0173.tif
new file mode 100644
index 00000000..6eb36412
Binary files /dev/null and b/tests/data/brain/image_0173.tif differ
diff --git a/tests/data/brain/image_0174.tif b/tests/data/brain/image_0174.tif
new file mode 100644
index 00000000..d29219b7
Binary files /dev/null and b/tests/data/brain/image_0174.tif differ
diff --git a/tests/data/brain/image_0175.tif b/tests/data/brain/image_0175.tif
new file mode 100644
index 00000000..6f8cff7b
Binary files /dev/null and b/tests/data/brain/image_0175.tif differ
diff --git a/tests/data/brain/image_0176.tif b/tests/data/brain/image_0176.tif
new file mode 100644
index 00000000..7058142c
Binary files /dev/null and b/tests/data/brain/image_0176.tif differ
diff --git a/tests/data/brain/image_0177.tif b/tests/data/brain/image_0177.tif
new file mode 100644
index 00000000..c6c11538
Binary files /dev/null and b/tests/data/brain/image_0177.tif differ
diff --git a/tests/data/brain/image_0178.tif b/tests/data/brain/image_0178.tif
new file mode 100644
index 00000000..791e8e03
Binary files /dev/null and b/tests/data/brain/image_0178.tif differ
diff --git a/tests/data/brain/image_0179.tif b/tests/data/brain/image_0179.tif
new file mode 100644
index 00000000..17a66bad
Binary files /dev/null and b/tests/data/brain/image_0179.tif differ
diff --git a/tests/data/brain/image_0180.tif b/tests/data/brain/image_0180.tif
new file mode 100644
index 00000000..5cebfff6
Binary files /dev/null and b/tests/data/brain/image_0180.tif differ
diff --git a/tests/data/brain/image_0181.tif b/tests/data/brain/image_0181.tif
new file mode 100644
index 00000000..f93e0aab
Binary files /dev/null and b/tests/data/brain/image_0181.tif differ
diff --git a/tests/data/brain/image_0182.tif b/tests/data/brain/image_0182.tif
new file mode 100644
index 00000000..cebca3bb
Binary files /dev/null and b/tests/data/brain/image_0182.tif differ
diff --git a/tests/data/brain/image_0183.tif b/tests/data/brain/image_0183.tif
new file mode 100644
index 00000000..bc7f033e
Binary files /dev/null and b/tests/data/brain/image_0183.tif differ
diff --git a/tests/data/brain/image_0184.tif b/tests/data/brain/image_0184.tif
new file mode 100644
index 00000000..a50dcef3
Binary files /dev/null and b/tests/data/brain/image_0184.tif differ
diff --git a/tests/data/brain/image_0185.tif b/tests/data/brain/image_0185.tif
new file mode 100644
index 00000000..3dd1e4e1
Binary files /dev/null and b/tests/data/brain/image_0185.tif differ
diff --git a/tests/data/brain/image_0186.tif b/tests/data/brain/image_0186.tif
new file mode 100644
index 00000000..1170e2a3
Binary files /dev/null and b/tests/data/brain/image_0186.tif differ
diff --git a/tests/data/brain/image_0187.tif b/tests/data/brain/image_0187.tif
new file mode 100644
index 00000000..1fc2037b
Binary files /dev/null and b/tests/data/brain/image_0187.tif differ
diff --git a/tests/data/brain/image_0188.tif b/tests/data/brain/image_0188.tif
new file mode 100644
index 00000000..ef569541
Binary files /dev/null and b/tests/data/brain/image_0188.tif differ
diff --git a/tests/data/brain/image_0189.tif b/tests/data/brain/image_0189.tif
new file mode 100644
index 00000000..3bc4d437
Binary files /dev/null and b/tests/data/brain/image_0189.tif differ
diff --git a/tests/data/brain/image_0190.tif b/tests/data/brain/image_0190.tif
new file mode 100644
index 00000000..be38ddfc
Binary files /dev/null and b/tests/data/brain/image_0190.tif differ
diff --git a/tests/data/brain/image_0191.tif b/tests/data/brain/image_0191.tif
new file mode 100644
index 00000000..05cae462
Binary files /dev/null and b/tests/data/brain/image_0191.tif differ
diff --git a/tests/data/brain/image_0192.tif b/tests/data/brain/image_0192.tif
new file mode 100644
index 00000000..dd367f09
Binary files /dev/null and b/tests/data/brain/image_0192.tif differ
diff --git a/tests/data/brain/image_0193.tif b/tests/data/brain/image_0193.tif
new file mode 100644
index 00000000..28ddfbad
Binary files /dev/null and b/tests/data/brain/image_0193.tif differ
diff --git a/tests/data/brain/image_0194.tif b/tests/data/brain/image_0194.tif
new file mode 100644
index 00000000..b91eda0c
Binary files /dev/null and b/tests/data/brain/image_0194.tif differ
diff --git a/tests/data/brain/image_0195.tif b/tests/data/brain/image_0195.tif
new file mode 100644
index 00000000..ec0cf209
Binary files /dev/null and b/tests/data/brain/image_0195.tif differ
diff --git a/tests/data/brain/image_0196.tif b/tests/data/brain/image_0196.tif
new file mode 100644
index 00000000..3dd56dc3
Binary files /dev/null and b/tests/data/brain/image_0196.tif differ
diff --git a/tests/data/brain/image_0197.tif b/tests/data/brain/image_0197.tif
new file mode 100644
index 00000000..8973ed72
Binary files /dev/null and b/tests/data/brain/image_0197.tif differ
diff --git a/tests/data/brain/image_0198.tif b/tests/data/brain/image_0198.tif
new file mode 100644
index 00000000..816c0f2b
Binary files /dev/null and b/tests/data/brain/image_0198.tif differ
diff --git a/tests/data/brain/image_0199.tif b/tests/data/brain/image_0199.tif
new file mode 100644
index 00000000..3d121b84
Binary files /dev/null and b/tests/data/brain/image_0199.tif differ
diff --git a/tests/data/brain/image_0200.tif b/tests/data/brain/image_0200.tif
new file mode 100644
index 00000000..ab0b9c19
Binary files /dev/null and b/tests/data/brain/image_0200.tif differ
diff --git a/tests/data/brain/image_0201.tif b/tests/data/brain/image_0201.tif
new file mode 100644
index 00000000..c023b509
Binary files /dev/null and b/tests/data/brain/image_0201.tif differ
diff --git a/tests/data/brain/image_0202.tif b/tests/data/brain/image_0202.tif
new file mode 100644
index 00000000..4395906f
Binary files /dev/null and b/tests/data/brain/image_0202.tif differ
diff --git a/tests/data/brain/image_0203.tif b/tests/data/brain/image_0203.tif
new file mode 100644
index 00000000..9691b45f
Binary files /dev/null and b/tests/data/brain/image_0203.tif differ
diff --git a/tests/data/brain/image_0204.tif b/tests/data/brain/image_0204.tif
new file mode 100644
index 00000000..6b24f9b1
Binary files /dev/null and b/tests/data/brain/image_0204.tif differ
diff --git a/tests/data/brain/image_0205.tif b/tests/data/brain/image_0205.tif
new file mode 100644
index 00000000..71e2304e
Binary files /dev/null and b/tests/data/brain/image_0205.tif differ
diff --git a/tests/data/brain/image_0206.tif b/tests/data/brain/image_0206.tif
new file mode 100644
index 00000000..1b2586ca
Binary files /dev/null and b/tests/data/brain/image_0206.tif differ
diff --git a/tests/data/brain/image_0207.tif b/tests/data/brain/image_0207.tif
new file mode 100644
index 00000000..af1baf55
Binary files /dev/null and b/tests/data/brain/image_0207.tif differ
diff --git a/tests/data/brain/image_0208.tif b/tests/data/brain/image_0208.tif
new file mode 100644
index 00000000..d8af70de
Binary files /dev/null and b/tests/data/brain/image_0208.tif differ
diff --git a/tests/data/brain/image_0209.tif b/tests/data/brain/image_0209.tif
new file mode 100644
index 00000000..574da21e
Binary files /dev/null and b/tests/data/brain/image_0209.tif differ
diff --git a/tests/data/brain/image_0210.tif b/tests/data/brain/image_0210.tif
new file mode 100644
index 00000000..ae63cab9
Binary files /dev/null and b/tests/data/brain/image_0210.tif differ
diff --git a/tests/data/brain/image_0211.tif b/tests/data/brain/image_0211.tif
new file mode 100644
index 00000000..1be8c71f
Binary files /dev/null and b/tests/data/brain/image_0211.tif differ
diff --git a/tests/data/brain/image_0212.tif b/tests/data/brain/image_0212.tif
new file mode 100644
index 00000000..7bfe262c
Binary files /dev/null and b/tests/data/brain/image_0212.tif differ
diff --git a/tests/data/brain/image_0213.tif b/tests/data/brain/image_0213.tif
new file mode 100644
index 00000000..d2dff52b
Binary files /dev/null and b/tests/data/brain/image_0213.tif differ
diff --git a/tests/data/brain/image_0214.tif b/tests/data/brain/image_0214.tif
new file mode 100644
index 00000000..fd8cbb11
Binary files /dev/null and b/tests/data/brain/image_0214.tif differ
diff --git a/tests/data/brain/image_0215.tif b/tests/data/brain/image_0215.tif
new file mode 100644
index 00000000..105f0d14
Binary files /dev/null and b/tests/data/brain/image_0215.tif differ
diff --git a/tests/data/brain/image_0216.tif b/tests/data/brain/image_0216.tif
new file mode 100644
index 00000000..025206c1
Binary files /dev/null and b/tests/data/brain/image_0216.tif differ
diff --git a/tests/data/brain/image_0217.tif b/tests/data/brain/image_0217.tif
new file mode 100644
index 00000000..b99ad148
Binary files /dev/null and b/tests/data/brain/image_0217.tif differ
diff --git a/tests/data/brain/image_0218.tif b/tests/data/brain/image_0218.tif
new file mode 100644
index 00000000..af69b0c5
Binary files /dev/null and b/tests/data/brain/image_0218.tif differ
diff --git a/tests/data/brain/image_0219.tif b/tests/data/brain/image_0219.tif
new file mode 100644
index 00000000..923e1873
Binary files /dev/null and b/tests/data/brain/image_0219.tif differ
diff --git a/tests/data/brain/image_0220.tif b/tests/data/brain/image_0220.tif
new file mode 100644
index 00000000..286e7140
Binary files /dev/null and b/tests/data/brain/image_0220.tif differ
diff --git a/tests/data/brain/image_0221.tif b/tests/data/brain/image_0221.tif
new file mode 100644
index 00000000..d9d1f124
Binary files /dev/null and b/tests/data/brain/image_0221.tif differ
diff --git a/tests/data/brain/image_0222.tif b/tests/data/brain/image_0222.tif
new file mode 100644
index 00000000..a9f8ceb3
Binary files /dev/null and b/tests/data/brain/image_0222.tif differ
diff --git a/tests/data/brain/image_0223.tif b/tests/data/brain/image_0223.tif
new file mode 100644
index 00000000..59df46d2
Binary files /dev/null and b/tests/data/brain/image_0223.tif differ
diff --git a/tests/data/brain/image_0224.tif b/tests/data/brain/image_0224.tif
new file mode 100644
index 00000000..ce3df3ff
Binary files /dev/null and b/tests/data/brain/image_0224.tif differ
diff --git a/tests/data/brain/image_0225.tif b/tests/data/brain/image_0225.tif
new file mode 100644
index 00000000..42ababef
Binary files /dev/null and b/tests/data/brain/image_0225.tif differ
diff --git a/tests/data/brain/image_0226.tif b/tests/data/brain/image_0226.tif
new file mode 100644
index 00000000..117b39f8
Binary files /dev/null and b/tests/data/brain/image_0226.tif differ
diff --git a/tests/data/brain/image_0227.tif b/tests/data/brain/image_0227.tif
new file mode 100644
index 00000000..c6f3090d
Binary files /dev/null and b/tests/data/brain/image_0227.tif differ
diff --git a/tests/data/brain/image_0228.tif b/tests/data/brain/image_0228.tif
new file mode 100644
index 00000000..5775774d
Binary files /dev/null and b/tests/data/brain/image_0228.tif differ
diff --git a/tests/data/brain/image_0229.tif b/tests/data/brain/image_0229.tif
new file mode 100644
index 00000000..1623ccb3
Binary files /dev/null and b/tests/data/brain/image_0229.tif differ
diff --git a/tests/data/brain/image_0230.tif b/tests/data/brain/image_0230.tif
new file mode 100644
index 00000000..e9bc9b7f
Binary files /dev/null and b/tests/data/brain/image_0230.tif differ
diff --git a/tests/data/brain/image_0231.tif b/tests/data/brain/image_0231.tif
new file mode 100644
index 00000000..2a1ed26b
Binary files /dev/null and b/tests/data/brain/image_0231.tif differ
diff --git a/tests/data/brain/image_0232.tif b/tests/data/brain/image_0232.tif
new file mode 100644
index 00000000..9ec937c7
Binary files /dev/null and b/tests/data/brain/image_0232.tif differ
diff --git a/tests/data/brain/image_0233.tif b/tests/data/brain/image_0233.tif
new file mode 100644
index 00000000..acc7019c
Binary files /dev/null and b/tests/data/brain/image_0233.tif differ
diff --git a/tests/data/brain/image_0234.tif b/tests/data/brain/image_0234.tif
new file mode 100644
index 00000000..1bdba899
Binary files /dev/null and b/tests/data/brain/image_0234.tif differ
diff --git a/tests/data/brain/image_0235.tif b/tests/data/brain/image_0235.tif
new file mode 100644
index 00000000..769464c0
Binary files /dev/null and b/tests/data/brain/image_0235.tif differ
diff --git a/tests/data/brain/image_0236.tif b/tests/data/brain/image_0236.tif
new file mode 100644
index 00000000..2c091f57
Binary files /dev/null and b/tests/data/brain/image_0236.tif differ
diff --git a/tests/data/brain/image_0237.tif b/tests/data/brain/image_0237.tif
new file mode 100644
index 00000000..e530c4af
Binary files /dev/null and b/tests/data/brain/image_0237.tif differ
diff --git a/tests/data/brain/image_0238.tif b/tests/data/brain/image_0238.tif
new file mode 100644
index 00000000..80b4cfee
Binary files /dev/null and b/tests/data/brain/image_0238.tif differ
diff --git a/tests/data/brain/image_0239.tif b/tests/data/brain/image_0239.tif
new file mode 100644
index 00000000..e64cfe6a
Binary files /dev/null and b/tests/data/brain/image_0239.tif differ
diff --git a/tests/data/brain/image_0240.tif b/tests/data/brain/image_0240.tif
new file mode 100644
index 00000000..569cdcce
Binary files /dev/null and b/tests/data/brain/image_0240.tif differ
diff --git a/tests/data/brain/image_0241.tif b/tests/data/brain/image_0241.tif
new file mode 100644
index 00000000..ca4713b8
Binary files /dev/null and b/tests/data/brain/image_0241.tif differ
diff --git a/tests/data/brain/image_0242.tif b/tests/data/brain/image_0242.tif
new file mode 100644
index 00000000..18e06f43
Binary files /dev/null and b/tests/data/brain/image_0242.tif differ
diff --git a/tests/data/brain/image_0243.tif b/tests/data/brain/image_0243.tif
new file mode 100644
index 00000000..acfca8ab
Binary files /dev/null and b/tests/data/brain/image_0243.tif differ
diff --git a/tests/data/brain/image_0244.tif b/tests/data/brain/image_0244.tif
new file mode 100644
index 00000000..6c3ff2fe
Binary files /dev/null and b/tests/data/brain/image_0244.tif differ
diff --git a/tests/data/brain/image_0245.tif b/tests/data/brain/image_0245.tif
new file mode 100644
index 00000000..665ef575
Binary files /dev/null and b/tests/data/brain/image_0245.tif differ
diff --git a/tests/data/brain/image_0246.tif b/tests/data/brain/image_0246.tif
new file mode 100644
index 00000000..72d8bd88
Binary files /dev/null and b/tests/data/brain/image_0246.tif differ
diff --git a/tests/data/brain/image_0247.tif b/tests/data/brain/image_0247.tif
new file mode 100644
index 00000000..f4136899
Binary files /dev/null and b/tests/data/brain/image_0247.tif differ
diff --git a/tests/data/brain/image_0248.tif b/tests/data/brain/image_0248.tif
new file mode 100644
index 00000000..984098e1
Binary files /dev/null and b/tests/data/brain/image_0248.tif differ
diff --git a/tests/data/brain/image_0249.tif b/tests/data/brain/image_0249.tif
new file mode 100644
index 00000000..0d88b096
Binary files /dev/null and b/tests/data/brain/image_0249.tif differ
diff --git a/tests/data/brain/image_0250.tif b/tests/data/brain/image_0250.tif
new file mode 100644
index 00000000..3c621f6c
Binary files /dev/null and b/tests/data/brain/image_0250.tif differ
diff --git a/tests/data/brain/image_0251.tif b/tests/data/brain/image_0251.tif
new file mode 100644
index 00000000..f983bd25
Binary files /dev/null and b/tests/data/brain/image_0251.tif differ
diff --git a/tests/data/brain/image_0252.tif b/tests/data/brain/image_0252.tif
new file mode 100644
index 00000000..d92f266a
Binary files /dev/null and b/tests/data/brain/image_0252.tif differ
diff --git a/tests/data/brain/image_0253.tif b/tests/data/brain/image_0253.tif
new file mode 100644
index 00000000..db0034c8
Binary files /dev/null and b/tests/data/brain/image_0253.tif differ
diff --git a/tests/data/brain/image_0254.tif b/tests/data/brain/image_0254.tif
new file mode 100644
index 00000000..819f9f7f
Binary files /dev/null and b/tests/data/brain/image_0254.tif differ
diff --git a/tests/data/brain/image_0255.tif b/tests/data/brain/image_0255.tif
new file mode 100644
index 00000000..3dc92d36
Binary files /dev/null and b/tests/data/brain/image_0255.tif differ
diff --git a/tests/data/brain/image_0256.tif b/tests/data/brain/image_0256.tif
new file mode 100644
index 00000000..27e619fb
Binary files /dev/null and b/tests/data/brain/image_0256.tif differ
diff --git a/tests/data/brain/image_0257.tif b/tests/data/brain/image_0257.tif
new file mode 100644
index 00000000..c7ebcbc2
Binary files /dev/null and b/tests/data/brain/image_0257.tif differ
diff --git a/tests/data/brain/image_0258.tif b/tests/data/brain/image_0258.tif
new file mode 100644
index 00000000..bd7df19b
Binary files /dev/null and b/tests/data/brain/image_0258.tif differ
diff --git a/tests/data/brain/image_0259.tif b/tests/data/brain/image_0259.tif
new file mode 100644
index 00000000..99eb59ae
Binary files /dev/null and b/tests/data/brain/image_0259.tif differ
diff --git a/tests/data/brain/image_0260.tif b/tests/data/brain/image_0260.tif
new file mode 100644
index 00000000..195e9895
Binary files /dev/null and b/tests/data/brain/image_0260.tif differ
diff --git a/tests/data/brain/image_0261.tif b/tests/data/brain/image_0261.tif
new file mode 100644
index 00000000..1e1de754
Binary files /dev/null and b/tests/data/brain/image_0261.tif differ
diff --git a/tests/data/brain/image_0262.tif b/tests/data/brain/image_0262.tif
new file mode 100644
index 00000000..dea579c5
Binary files /dev/null and b/tests/data/brain/image_0262.tif differ
diff --git a/tests/data/brain/image_0263.tif b/tests/data/brain/image_0263.tif
new file mode 100644
index 00000000..ac0f32fa
Binary files /dev/null and b/tests/data/brain/image_0263.tif differ
diff --git a/tests/data/brain/image_0264.tif b/tests/data/brain/image_0264.tif
new file mode 100644
index 00000000..5879819f
Binary files /dev/null and b/tests/data/brain/image_0264.tif differ
diff --git a/tests/data/brain/image_0265.tif b/tests/data/brain/image_0265.tif
new file mode 100644
index 00000000..a920e777
Binary files /dev/null and b/tests/data/brain/image_0265.tif differ
diff --git a/tests/data/brain/image_0266.tif b/tests/data/brain/image_0266.tif
new file mode 100644
index 00000000..97beecbf
Binary files /dev/null and b/tests/data/brain/image_0266.tif differ
diff --git a/tests/data/brain/image_0267.tif b/tests/data/brain/image_0267.tif
new file mode 100644
index 00000000..80c46cdd
Binary files /dev/null and b/tests/data/brain/image_0267.tif differ
diff --git a/tests/data/brain/image_0268.tif b/tests/data/brain/image_0268.tif
new file mode 100644
index 00000000..fb01fa63
Binary files /dev/null and b/tests/data/brain/image_0268.tif differ
diff --git a/tests/data/brain/image_0269.tif b/tests/data/brain/image_0269.tif
new file mode 100644
index 00000000..d95b0215
Binary files /dev/null and b/tests/data/brain/image_0269.tif differ
diff --git a/tests/data/cells/cells.xml b/tests/data/cells/cells.xml
new file mode 100644
index 00000000..10df74ad
--- /dev/null
+++ b/tests/data/cells/cells.xml
@@ -0,0 +1,337 @@
+
+
+
+ placeholder.tif
+
+
+ 1
+
+ 1
+
+ 181
+ 2379
+ 1282
+
+
+ 2117
+ 133
+ 1276
+
+
+ 4082
+ 332
+ 1281
+
+
+ 2137
+ 134
+ 1286
+
+
+ 2129
+ 133
+ 1276
+
+
+ 2141
+ 134
+ 1278
+
+
+ 2130
+ 132
+ 1277
+
+
+ 2130
+ 133
+ 1278
+
+
+ 2133
+ 136
+ 1285
+
+
+ 2141
+ 136
+ 1297
+
+
+ 2162
+ 130
+ 1274
+
+
+ 2127
+ 1026
+ 1294
+
+
+ 1801
+ 336
+ 1290
+
+
+ 2176
+ 134
+ 1279
+
+
+ 2194
+ 134
+ 1280
+
+
+ 2664
+ 336
+ 1296
+
+
+ 2435
+ 343
+ 1290
+
+
+ 1266
+ 3427
+ 1294
+
+
+ 568
+ 2012
+ 1294
+
+
+ 2103
+ 138
+ 1285
+
+
+ 2098
+ 138
+ 1279
+
+
+ 2105
+ 140
+ 1289
+
+
+ 2208
+ 135
+ 1275
+
+
+ 2162
+ 134
+ 1286
+
+
+ 2221
+ 137
+ 1281
+
+
+ 2081
+ 142
+ 1284
+
+
+ 2230
+ 139
+ 1275
+
+
+ 2015
+ 1045
+ 1291
+
+
+ 2072
+ 141
+ 1273
+
+
+ 1413
+ 2045
+ 1294
+
+
+ 1950
+ 1048
+ 1294
+
+
+ 3340
+ 142
+ 1274
+
+
+ 2051
+ 142
+ 1274
+
+
+ 3362
+ 145
+ 1274
+
+
+ 2046
+ 150
+ 1283
+
+
+ 2046
+ 151
+ 1287
+
+
+ 2046
+ 151
+ 1290
+
+
+ 2047
+ 151
+ 1294
+
+
+ 2032
+ 146
+ 1272
+
+
+ 2923
+ 147
+ 1275
+
+
+ 2064
+ 147
+ 1275
+
+
+ 2267
+ 147
+ 1274
+
+
+ 2297
+ 1068
+ 1292
+
+
+ 2004
+ 149
+ 1273
+
+
+ 2356
+ 521
+ 1288
+
+
+ 2357
+ 522
+ 1286
+
+
+ 2359
+ 523
+ 1287
+
+
+ 2360
+ 523
+ 1289
+
+
+ 2365
+ 525
+ 1298
+
+
+ 2612
+ 520
+ 1274
+
+
+ 2623
+ 520
+ 1274
+
+
+ 2657
+ 521
+ 1273
+
+
+ 2906
+ 522
+ 1275
+
+
+ 2165
+ 524
+ 1275
+
+
+ 2685
+ 522
+ 1278
+
+
+ 2892
+ 526
+ 1286
+
+
+ 2896
+ 525
+ 1278
+
+
+ 2894
+ 525
+ 1288
+
+
+ 2901
+ 526
+ 1295
+
+
+ 2270
+ 524
+ 1279
+
+
+ 1924
+ 974
+ 1282
+
+
+ 2505
+ 522
+ 1275
+
+
+ 2531
+ 527
+ 1276
+
+
+ 2542
+ 524
+ 1275
+
+
+ 2536
+ 523
+ 1286
+
+
+
+
diff --git a/tests/data/config/__init__.py b/tests/data/config/__init__.py
new file mode 100644
index 00000000..e69de29b
diff --git a/tests/data/config/test.conf b/tests/data/config/test.conf
new file mode 100644
index 00000000..94262784
--- /dev/null
+++ b/tests/data/config/test.conf
@@ -0,0 +1,18 @@
+[model]
+ model_path = ''
+ model_type = '50-layer'
+
+[atlas]
+ base_folder = ''
+ atlas_name = 'annotations.nii'
+ brain_name = 'brain_filtered.nii'
+ hemispheres_name = 'hemispheres.nii'
+ structures_name = 'structures.csv'
+ orientation = 'horizontal'
+ left_hemisphere_value = 2
+ right_hemisphere_value = 1
+
+ [[pixel_size]]
+ x = 100
+ y = 100
+ z = 100
diff --git a/tests/data/cube_extract/cells.xml b/tests/data/cube_extract/cells.xml
new file mode 100644
index 00000000..c8d5deba
--- /dev/null
+++ b/tests/data/cube_extract/cells.xml
@@ -0,0 +1,22 @@
+
+
+
+ placeholder.tif
+
+
+ 1
+
+ 1
+
+ 340
+ 1004
+ 15
+
+
+ 392
+ 522
+ 10
+
+
+
+
diff --git a/tests/data/cube_extract/cubes/pCellz10y522x392Ch0.tif b/tests/data/cube_extract/cubes/pCellz10y522x392Ch0.tif
new file mode 100644
index 00000000..d5c60fae
Binary files /dev/null and b/tests/data/cube_extract/cubes/pCellz10y522x392Ch0.tif differ
diff --git a/tests/data/cube_extract/cubes/pCellz10y522x392Ch1.tif b/tests/data/cube_extract/cubes/pCellz10y522x392Ch1.tif
new file mode 100644
index 00000000..1df64a6d
Binary files /dev/null and b/tests/data/cube_extract/cubes/pCellz10y522x392Ch1.tif differ
diff --git a/tests/data/cube_extract/cubes/pCellz15y1004x340Ch0.tif b/tests/data/cube_extract/cubes/pCellz15y1004x340Ch0.tif
new file mode 100644
index 00000000..08cb9dec
Binary files /dev/null and b/tests/data/cube_extract/cubes/pCellz15y1004x340Ch0.tif differ
diff --git a/tests/data/cube_extract/cubes/pCellz15y1004x340Ch1.tif b/tests/data/cube_extract/cubes/pCellz15y1004x340Ch1.tif
new file mode 100644
index 00000000..1cc9d859
Binary files /dev/null and b/tests/data/cube_extract/cubes/pCellz15y1004x340Ch1.tif differ
diff --git a/tests/data/cube_extract/cubes_scale/pCellz10y522x392Ch0.tif b/tests/data/cube_extract/cubes_scale/pCellz10y522x392Ch0.tif
new file mode 100644
index 00000000..bb5e0a97
Binary files /dev/null and b/tests/data/cube_extract/cubes_scale/pCellz10y522x392Ch0.tif differ
diff --git a/tests/data/cube_extract/cubes_scale/pCellz10y522x392Ch1.tif b/tests/data/cube_extract/cubes_scale/pCellz10y522x392Ch1.tif
new file mode 100644
index 00000000..6bef30b2
Binary files /dev/null and b/tests/data/cube_extract/cubes_scale/pCellz10y522x392Ch1.tif differ
diff --git a/tests/data/cube_extract/cubes_scale/pCellz15y1004x340Ch0.tif b/tests/data/cube_extract/cubes_scale/pCellz15y1004x340Ch0.tif
new file mode 100644
index 00000000..a0cabc2f
Binary files /dev/null and b/tests/data/cube_extract/cubes_scale/pCellz15y1004x340Ch0.tif differ
diff --git a/tests/data/cube_extract/cubes_scale/pCellz15y1004x340Ch1.tif b/tests/data/cube_extract/cubes_scale/pCellz15y1004x340Ch1.tif
new file mode 100644
index 00000000..1b9acc69
Binary files /dev/null and b/tests/data/cube_extract/cubes_scale/pCellz15y1004x340Ch1.tif differ
diff --git a/tests/data/image_processing/despeckled.tif b/tests/data/image_processing/despeckled.tif
new file mode 100644
index 00000000..90969162
Binary files /dev/null and b/tests/data/image_processing/despeckled.tif differ
diff --git a/tests/data/image_processing/flatfield.tif b/tests/data/image_processing/flatfield.tif
new file mode 100644
index 00000000..7df7f147
Binary files /dev/null and b/tests/data/image_processing/flatfield.tif differ
diff --git a/tests/data/integration/detection/cell_classification.xml b/tests/data/integration/detection/cell_classification.xml
new file mode 100644
index 00000000..ac7b5b73
--- /dev/null
+++ b/tests/data/integration/detection/cell_classification.xml
@@ -0,0 +1,292 @@
+
+
+
+ placeholder.tif
+
+
+ 1
+
+ 2
+
+ 132
+ 308
+ 10
+
+
+ 216
+ 492
+ 14
+
+
+ 223
+ 133
+ 14
+
+
+ 195
+ 209
+ 14
+
+
+ 240
+ 253
+ 14
+
+
+ 159
+ 318
+ 14
+
+
+ 208
+ 338
+ 14
+
+
+ 317
+ 125
+ 12
+
+
+ 119
+ 165
+ 12
+
+
+ 205
+ 244
+ 12
+
+
+ 125
+ 264
+ 12
+
+
+ 274
+ 310
+ 12
+
+
+ 122
+ 338
+ 12
+
+
+ 630
+ 359
+ 12
+
+
+ 163
+ 403
+ 12
+
+
+ 185
+ 296
+ 12
+
+
+ 171
+ 297
+ 12
+
+
+ 103
+ 303
+ 12
+
+
+ 187
+ 153
+ 13
+
+
+ 207
+ 443
+ 13
+
+
+ 156
+ 400
+ 13
+
+
+ 128
+ 421
+ 13
+
+
+ 142
+ 159
+ 13
+
+
+ 196
+ 462
+ 13
+
+
+ 319
+ 211
+ 11
+
+
+ 49
+ 482
+ 11
+
+
+ 304
+ 110
+ 11
+
+
+ 141
+ 159
+ 11
+
+
+ 236
+ 212
+ 15
+
+
+ 223
+ 269
+ 15
+
+
+ 165
+ 293
+ 15
+
+
+ 138
+ 296
+ 15
+
+
+ 188
+ 333
+ 15
+
+
+ 214
+ 352
+ 15
+
+
+ 236
+ 358
+ 15
+
+
+ 214
+ 462
+ 15
+
+
+ 234
+ 233
+ 15
+
+
+ 189
+ 252
+ 15
+
+
+ 145
+ 272
+ 15
+
+
+ 57
+ 296
+ 15
+
+
+ 142
+ 421
+ 15
+
+
+ 98
+ 343
+ 15
+
+
+ 213
+ 277
+ 17
+
+
+ 597
+ 254
+ 17
+
+
+ 415
+ 143
+ 17
+
+
+ 198
+ 331
+ 17
+
+
+ 129
+ 422
+ 17
+
+
+ 165
+ 305
+ 19
+
+
+ 183
+ 424
+ 16
+
+
+ 203
+ 381
+ 16
+
+
+ 278
+ 237
+ 16
+
+
+ 187
+ 295
+ 16
+
+
+ 158
+ 400
+ 16
+
+
+ 99
+ 342
+ 18
+
+
+ 198
+ 429
+ 18
+
+
+ 240
+ 468
+ 18
+
+
+
+
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00000.tif b/tests/data/integration/detection/crop_planes/ch0/ch00000.tif
new file mode 100644
index 00000000..abca45a0
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00000.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00001.tif b/tests/data/integration/detection/crop_planes/ch0/ch00001.tif
new file mode 100644
index 00000000..7f59b3d0
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00001.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00002.tif b/tests/data/integration/detection/crop_planes/ch0/ch00002.tif
new file mode 100644
index 00000000..c2b4eb96
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00002.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00003.tif b/tests/data/integration/detection/crop_planes/ch0/ch00003.tif
new file mode 100644
index 00000000..eec563fe
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00003.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00004.tif b/tests/data/integration/detection/crop_planes/ch0/ch00004.tif
new file mode 100644
index 00000000..11600d10
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00004.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00005.tif b/tests/data/integration/detection/crop_planes/ch0/ch00005.tif
new file mode 100644
index 00000000..e6ff786d
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00005.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00006.tif b/tests/data/integration/detection/crop_planes/ch0/ch00006.tif
new file mode 100644
index 00000000..d17b219a
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00006.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00007.tif b/tests/data/integration/detection/crop_planes/ch0/ch00007.tif
new file mode 100644
index 00000000..93757586
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00007.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00008.tif b/tests/data/integration/detection/crop_planes/ch0/ch00008.tif
new file mode 100644
index 00000000..879bd53a
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00008.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00009.tif b/tests/data/integration/detection/crop_planes/ch0/ch00009.tif
new file mode 100644
index 00000000..1492fd49
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00009.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00010.tif b/tests/data/integration/detection/crop_planes/ch0/ch00010.tif
new file mode 100644
index 00000000..9eccc8c9
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00010.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00011.tif b/tests/data/integration/detection/crop_planes/ch0/ch00011.tif
new file mode 100644
index 00000000..a116ea35
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00011.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00012.tif b/tests/data/integration/detection/crop_planes/ch0/ch00012.tif
new file mode 100644
index 00000000..7dd73e6c
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00012.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00013.tif b/tests/data/integration/detection/crop_planes/ch0/ch00013.tif
new file mode 100644
index 00000000..768d0fb3
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00013.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00014.tif b/tests/data/integration/detection/crop_planes/ch0/ch00014.tif
new file mode 100644
index 00000000..c8f9df08
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00014.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00015.tif b/tests/data/integration/detection/crop_planes/ch0/ch00015.tif
new file mode 100644
index 00000000..5a128b1e
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00015.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00016.tif b/tests/data/integration/detection/crop_planes/ch0/ch00016.tif
new file mode 100644
index 00000000..8c36d48e
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00016.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00017.tif b/tests/data/integration/detection/crop_planes/ch0/ch00017.tif
new file mode 100644
index 00000000..e72594f3
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00017.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00018.tif b/tests/data/integration/detection/crop_planes/ch0/ch00018.tif
new file mode 100644
index 00000000..90091d5b
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00018.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00019.tif b/tests/data/integration/detection/crop_planes/ch0/ch00019.tif
new file mode 100644
index 00000000..1626c60c
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00019.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00020.tif b/tests/data/integration/detection/crop_planes/ch0/ch00020.tif
new file mode 100644
index 00000000..6cef6e18
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00020.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00021.tif b/tests/data/integration/detection/crop_planes/ch0/ch00021.tif
new file mode 100644
index 00000000..f17445dc
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00021.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00022.tif b/tests/data/integration/detection/crop_planes/ch0/ch00022.tif
new file mode 100644
index 00000000..71ecb688
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00022.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00023.tif b/tests/data/integration/detection/crop_planes/ch0/ch00023.tif
new file mode 100644
index 00000000..a9c3cfed
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00023.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00024.tif b/tests/data/integration/detection/crop_planes/ch0/ch00024.tif
new file mode 100644
index 00000000..5bab6d10
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00024.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00025.tif b/tests/data/integration/detection/crop_planes/ch0/ch00025.tif
new file mode 100644
index 00000000..082d90bc
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00025.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00026.tif b/tests/data/integration/detection/crop_planes/ch0/ch00026.tif
new file mode 100644
index 00000000..390241c5
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00026.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00027.tif b/tests/data/integration/detection/crop_planes/ch0/ch00027.tif
new file mode 100644
index 00000000..2f293dec
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00027.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00028.tif b/tests/data/integration/detection/crop_planes/ch0/ch00028.tif
new file mode 100644
index 00000000..effeb8a2
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00028.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch0/ch00029.tif b/tests/data/integration/detection/crop_planes/ch0/ch00029.tif
new file mode 100644
index 00000000..4cfec35b
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch0/ch00029.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10000.tif b/tests/data/integration/detection/crop_planes/ch1/ch10000.tif
new file mode 100644
index 00000000..d4a2398f
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10000.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10001.tif b/tests/data/integration/detection/crop_planes/ch1/ch10001.tif
new file mode 100644
index 00000000..801ca48c
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10001.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10002.tif b/tests/data/integration/detection/crop_planes/ch1/ch10002.tif
new file mode 100644
index 00000000..74f9593b
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10002.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10003.tif b/tests/data/integration/detection/crop_planes/ch1/ch10003.tif
new file mode 100644
index 00000000..21a5a049
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10003.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10004.tif b/tests/data/integration/detection/crop_planes/ch1/ch10004.tif
new file mode 100644
index 00000000..15576026
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10004.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10005.tif b/tests/data/integration/detection/crop_planes/ch1/ch10005.tif
new file mode 100644
index 00000000..53ac6f78
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10005.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10006.tif b/tests/data/integration/detection/crop_planes/ch1/ch10006.tif
new file mode 100644
index 00000000..a7510b68
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10006.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10007.tif b/tests/data/integration/detection/crop_planes/ch1/ch10007.tif
new file mode 100644
index 00000000..8dc0836f
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10007.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10008.tif b/tests/data/integration/detection/crop_planes/ch1/ch10008.tif
new file mode 100644
index 00000000..8f9531b5
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10008.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10009.tif b/tests/data/integration/detection/crop_planes/ch1/ch10009.tif
new file mode 100644
index 00000000..f4d89f97
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10009.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10010.tif b/tests/data/integration/detection/crop_planes/ch1/ch10010.tif
new file mode 100644
index 00000000..fd995d0e
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10010.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10011.tif b/tests/data/integration/detection/crop_planes/ch1/ch10011.tif
new file mode 100644
index 00000000..059d274a
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10011.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10012.tif b/tests/data/integration/detection/crop_planes/ch1/ch10012.tif
new file mode 100644
index 00000000..045c9373
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10012.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10013.tif b/tests/data/integration/detection/crop_planes/ch1/ch10013.tif
new file mode 100644
index 00000000..546b0793
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10013.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10014.tif b/tests/data/integration/detection/crop_planes/ch1/ch10014.tif
new file mode 100644
index 00000000..72732ff2
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10014.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10015.tif b/tests/data/integration/detection/crop_planes/ch1/ch10015.tif
new file mode 100644
index 00000000..08831473
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10015.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10016.tif b/tests/data/integration/detection/crop_planes/ch1/ch10016.tif
new file mode 100644
index 00000000..d35b957c
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10016.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10017.tif b/tests/data/integration/detection/crop_planes/ch1/ch10017.tif
new file mode 100644
index 00000000..c21d7ac9
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10017.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10018.tif b/tests/data/integration/detection/crop_planes/ch1/ch10018.tif
new file mode 100644
index 00000000..47b97660
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10018.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10019.tif b/tests/data/integration/detection/crop_planes/ch1/ch10019.tif
new file mode 100644
index 00000000..2a7577e0
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10019.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10020.tif b/tests/data/integration/detection/crop_planes/ch1/ch10020.tif
new file mode 100644
index 00000000..99530a9f
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10020.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10021.tif b/tests/data/integration/detection/crop_planes/ch1/ch10021.tif
new file mode 100644
index 00000000..243309b0
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10021.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10022.tif b/tests/data/integration/detection/crop_planes/ch1/ch10022.tif
new file mode 100644
index 00000000..09d97b35
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10022.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10023.tif b/tests/data/integration/detection/crop_planes/ch1/ch10023.tif
new file mode 100644
index 00000000..9d87ac4f
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10023.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10024.tif b/tests/data/integration/detection/crop_planes/ch1/ch10024.tif
new file mode 100644
index 00000000..38fa3af3
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10024.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10025.tif b/tests/data/integration/detection/crop_planes/ch1/ch10025.tif
new file mode 100644
index 00000000..05634da5
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10025.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10026.tif b/tests/data/integration/detection/crop_planes/ch1/ch10026.tif
new file mode 100644
index 00000000..d9be83e7
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10026.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10027.tif b/tests/data/integration/detection/crop_planes/ch1/ch10027.tif
new file mode 100644
index 00000000..5daacc7c
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10027.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10028.tif b/tests/data/integration/detection/crop_planes/ch1/ch10028.tif
new file mode 100644
index 00000000..146ce30e
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10028.tif differ
diff --git a/tests/data/integration/detection/crop_planes/ch1/ch10029.tif b/tests/data/integration/detection/crop_planes/ch1/ch10029.tif
new file mode 100644
index 00000000..1a9409ca
Binary files /dev/null and b/tests/data/integration/detection/crop_planes/ch1/ch10029.tif differ
diff --git a/tests/data/integration/training/cells/pCellz1132y413x1896Ch0.tif b/tests/data/integration/training/cells/pCellz1132y413x1896Ch0.tif
new file mode 100644
index 00000000..682b0c1a
Binary files /dev/null and b/tests/data/integration/training/cells/pCellz1132y413x1896Ch0.tif differ
diff --git a/tests/data/integration/training/cells/pCellz1132y413x1896Ch1.tif b/tests/data/integration/training/cells/pCellz1132y413x1896Ch1.tif
new file mode 100644
index 00000000..d3454156
Binary files /dev/null and b/tests/data/integration/training/cells/pCellz1132y413x1896Ch1.tif differ
diff --git a/tests/data/integration/training/cells/pCellz1138y502x1662Ch0.tif b/tests/data/integration/training/cells/pCellz1138y502x1662Ch0.tif
new file mode 100644
index 00000000..a7dd151f
Binary files /dev/null and b/tests/data/integration/training/cells/pCellz1138y502x1662Ch0.tif differ
diff --git a/tests/data/integration/training/cells/pCellz1138y502x1662Ch1.tif b/tests/data/integration/training/cells/pCellz1138y502x1662Ch1.tif
new file mode 100644
index 00000000..c55cad07
Binary files /dev/null and b/tests/data/integration/training/cells/pCellz1138y502x1662Ch1.tif differ
diff --git a/tests/data/integration/training/non_cells/pCellz1137y278x1616Ch0.tif b/tests/data/integration/training/non_cells/pCellz1137y278x1616Ch0.tif
new file mode 100644
index 00000000..0dc4da1d
Binary files /dev/null and b/tests/data/integration/training/non_cells/pCellz1137y278x1616Ch0.tif differ
diff --git a/tests/data/integration/training/non_cells/pCellz1137y278x1616Ch1.tif b/tests/data/integration/training/non_cells/pCellz1137y278x1616Ch1.tif
new file mode 100644
index 00000000..ed817eab
Binary files /dev/null and b/tests/data/integration/training/non_cells/pCellz1137y278x1616Ch1.tif differ
diff --git a/tests/data/integration/training/non_cells/pCellz1137y426x1996Ch0.tif b/tests/data/integration/training/non_cells/pCellz1137y426x1996Ch0.tif
new file mode 100644
index 00000000..48a94570
Binary files /dev/null and b/tests/data/integration/training/non_cells/pCellz1137y426x1996Ch0.tif differ
diff --git a/tests/data/integration/training/non_cells/pCellz1137y426x1996Ch1.tif b/tests/data/integration/training/non_cells/pCellz1137y426x1996Ch1.tif
new file mode 100644
index 00000000..771cde8e
Binary files /dev/null and b/tests/data/integration/training/non_cells/pCellz1137y426x1996Ch1.tif differ
diff --git a/tests/data/integration/training/training.yml b/tests/data/integration/training/training.yml
new file mode 100644
index 00000000..90e0a634
--- /dev/null
+++ b/tests/data/integration/training/training.yml
@@ -0,0 +1,11 @@
+data:
+- bg_channel: 1
+ cell_def: ''
+ cube_dir: tests/data/integration/training/cells
+ signal_channel: 0
+ type: cell
+- bg_channel: 1
+ cell_def: ''
+ cube_dir: tests/data/integration/training/cells
+ signal_channel: 0
+ type: no_cell
diff --git a/tests/data/registration_output/Linux/boundaries.tiff b/tests/data/registration_output/Linux/boundaries.tiff
new file mode 100644
index 00000000..76754cc1
Binary files /dev/null and b/tests/data/registration_output/Linux/boundaries.tiff differ
diff --git a/tests/data/registration_output/Linux/deformation_field_0.tiff b/tests/data/registration_output/Linux/deformation_field_0.tiff
new file mode 100644
index 00000000..3947ceef
Binary files /dev/null and b/tests/data/registration_output/Linux/deformation_field_0.tiff differ
diff --git a/tests/data/registration_output/Linux/deformation_field_1.tiff b/tests/data/registration_output/Linux/deformation_field_1.tiff
new file mode 100644
index 00000000..793184bd
Binary files /dev/null and b/tests/data/registration_output/Linux/deformation_field_1.tiff differ
diff --git a/tests/data/registration_output/Linux/deformation_field_2.tiff b/tests/data/registration_output/Linux/deformation_field_2.tiff
new file mode 100644
index 00000000..62c97cdf
Binary files /dev/null and b/tests/data/registration_output/Linux/deformation_field_2.tiff differ
diff --git a/tests/data/registration_output/Linux/downsampled.tiff b/tests/data/registration_output/Linux/downsampled.tiff
new file mode 100644
index 00000000..68270cb2
Binary files /dev/null and b/tests/data/registration_output/Linux/downsampled.tiff differ
diff --git a/tests/data/registration_output/Linux/downsampled_channel_0.tiff b/tests/data/registration_output/Linux/downsampled_channel_0.tiff
new file mode 100644
index 00000000..68270cb2
Binary files /dev/null and b/tests/data/registration_output/Linux/downsampled_channel_0.tiff differ
diff --git a/tests/data/registration_output/Linux/downsampled_standard.tiff b/tests/data/registration_output/Linux/downsampled_standard.tiff
new file mode 100644
index 00000000..60ea2df9
Binary files /dev/null and b/tests/data/registration_output/Linux/downsampled_standard.tiff differ
diff --git a/tests/data/registration_output/Linux/downsampled_standard_channel_0.tiff b/tests/data/registration_output/Linux/downsampled_standard_channel_0.tiff
new file mode 100644
index 00000000..60ea2df9
Binary files /dev/null and b/tests/data/registration_output/Linux/downsampled_standard_channel_0.tiff differ
diff --git a/tests/data/registration_output/Linux/registered_atlas.tiff b/tests/data/registration_output/Linux/registered_atlas.tiff
new file mode 100644
index 00000000..98431721
Binary files /dev/null and b/tests/data/registration_output/Linux/registered_atlas.tiff differ
diff --git a/tests/data/registration_output/Linux/registered_hemispheres.tiff b/tests/data/registration_output/Linux/registered_hemispheres.tiff
new file mode 100644
index 00000000..6a4af26c
Binary files /dev/null and b/tests/data/registration_output/Linux/registered_hemispheres.tiff differ
diff --git a/tests/data/registration_output/Linux/volumes.csv b/tests/data/registration_output/Linux/volumes.csv
new file mode 100644
index 00000000..91416932
--- /dev/null
+++ b/tests/data/registration_output/Linux/volumes.csv
@@ -0,0 +1,663 @@
+structure_name,left_volume_mm3,right_volume_mm3,total_volume_mm3
+"Tuberomammillary nucleus, ventral part",0.057,0.056,0.113
+"Primary somatosensory area, mouth, layer 6b",0.06,0.061,0.121
+internal capsule,1.025,1.049,2.074
+Principal sensory nucleus of the trigeminal,0.589,0.5630000000000001,1.1520000000000001
+"Primary somatosensory area, trunk, layer 6a",0.138,0.114,0.252
+"Superior colliculus, motor related, intermediate gray layer",0.966,1.027,1.9929999999999999
+Interfascicular nucleus raphe,0.033,0.058,0.091
+Parataenial nucleus,0.11800000000000001,0.122,0.24
+"Superior colliculus, motor related, intermediate white layer",0.99,1.073,2.0629999999999997
+Induseum griseum,0.032,0.07200000000000001,0.10400000000000001
+"Entorhinal area, lateral part, layer 2",0.803,0.8,1.6030000000000002
+Anterior amygdalar area,0.23700000000000002,0.242,0.479
+"Superior colliculus, motor related, deep gray layer",0.5690000000000001,0.596,1.165
+Intergeniculate leaflet of the lateral geniculate complex,0.036000000000000004,0.035,0.07100000000000001
+"Entorhinal area, lateral part, layer 6a",0.545,0.528,1.073
+"Primary visual area, layer 6a",0.527,0.522,1.049
+Oculomotor nucleus,0.013000000000000001,0.014,0.027000000000000003
+"Gustatory areas, layer 1",0.1,0.101,0.201
+Paraventricular hypothalamic nucleus,0.089,0.098,0.187
+"posteromedial visual area, layer 2/3",0.14100000000000001,0.154,0.29500000000000004
+"Superior colliculus, motor related, deep white layer",0.162,0.158,0.32
+Precommissural nucleus,0.098,0.101,0.199
+"Entorhinal area, lateral part, layer 3",0.673,0.649,1.322
+medial forebrain bundle,0.03,0.024,0.054
+Nucleus accumbens,2.236,2.226,4.462
+Medial terminal nucleus of the accessory optic tract,0.026000000000000002,0.025,0.051000000000000004
+Intermediodorsal nucleus of the thalamus,0.045,0.137,0.182
+medial longitudinal fascicle,0.041,0.046,0.087
+"Paraventricular hypothalamic nucleus, descending division",0.075,0.065,0.14
+Anterodorsal nucleus,0.083,0.075,0.158
+Lateral terminal nucleus of the accessory optic tract,0.01,0.009000000000000001,0.019000000000000003
+Interstitial nucleus of Cajal,0.049,0.047,0.096
+"Frontal pole, layer 1",0.112,0.123,0.235
+Anterodorsal preoptic nucleus,0.053,0.053,0.106
+"Lateral visual area, layer 6a",0.10200000000000001,0.101,0.203
+Dorsal terminal nucleus of the accessory optic tract,0.006,0.006,0.012
+middle cerebellar peduncle,0.52,0.518,1.038
+lateral ventricle,1.002,1.114,2.116
+Inferior olivary complex,0.23800000000000002,0.256,0.494
+"Prelimbic area, layer 6a",0.212,0.23700000000000002,0.449
+Anterior hypothalamic nucleus,0.388,0.375,0.763
+Interposed nucleus,0.396,0.428,0.8240000000000001
+motor root of the trigeminal nerve,0.031,0.033,0.064
+Dorsal cochlear nucleus,0.302,0.317,0.619
+"Temporal association areas, layer 1",0.254,0.252,0.506
+subependymal zone,0.063,0.066,0.129
+Interpeduncular nucleus,0.003,0.006,0.009000000000000001
+Ventral cochlear nucleus,0.531,0.537,1.068
+nigrostriatal tract,0.054,0.064,0.118
+"Superior olivary complex, medial part",0.10200000000000001,0.10300000000000001,0.20500000000000002
+Inferior salivatory nucleus,0.005,0.005,0.01
+choroid plexus,0.638,0.775,1.413
+"Primary somatosensory area, lower limb, layer 2/3",0.357,0.34600000000000003,0.7030000000000001
+"Superior olivary complex, lateral part",0.17300000000000001,0.176,0.349
+Trochlear nucleus,0.003,0.003,0.006
+optic chiasm,0.151,0.17500000000000002,0.326
+"Periventricular hypothalamic nucleus, intermediate part",0.029,0.121,0.15
+"Agranular insular area, posterior part, layer 1",0.215,0.221,0.436
+"Lateral visual area, layer 6b",0.017,0.017,0.034
+"Superior olivary complex, periolivary region",0.196,0.192,0.388
+Koelliker-Fuse subnucleus,0.08600000000000001,0.08600000000000001,0.17200000000000001
+optic tract,0.34400000000000003,0.367,0.7110000000000001
+"Periventricular hypothalamic nucleus, posterior part",0.058,0.064,0.122
+Midbrain reticular nucleus,2.6390000000000002,2.589,5.228
+third ventricle,0.26,0.796,1.056
+Lateral amygdalar nucleus,0.417,0.43,0.847
+"Prelimbic area, layer 6b",0.011,0.008,0.019
+"Periventricular hypothalamic nucleus, preoptic part",0.022,0.096,0.118
+Intermediate reticular nucleus,1.397,1.418,2.815
+"Entorhinal area, lateral part, layer 5",0.709,0.713,1.422
+cerebral aqueduct,0.156,0.266,0.42200000000000004
+"Nucleus ambiguus, ventral division",0.008,0.009000000000000001,0.017
+fourth ventricle,0.227,0.299,0.526
+Pontine reticular nucleus,1.192,1.175,2.367
+Locus ceruleus,0.007,0.006,0.013000000000000001
+"Gustatory areas, layer 4",0.07200000000000001,0.069,0.14100000000000001
+Paraventricular nucleus of the thalamus,0.17,0.273,0.44300000000000006
+lateral recess,0.23800000000000002,0.23800000000000002,0.47600000000000003
+Lateral dorsal nucleus of thalamus,0.528,0.486,1.014
+"Dorsal auditory area, layer 6a",0.107,0.098,0.20500000000000002
+posterior commissure,0.019,0.03,0.049
+Anterior olfactory nucleus,2.448,2.497,4.945
+Laterodorsal tegmental nucleus,0.089,0.10300000000000001,0.192
+"Agranular insular area, posterior part, layer 2/3",0.397,0.402,0.799
+Nucleus prepositus,0.112,0.128,0.24
+"Prelimbic area, layer 1",0.209,0.328,0.537
+Retrochiasmatic area,0.077,0.068,0.14500000000000002
+Nucleus of Roller,0.014,0.017,0.031
+Ventral part of the lateral geniculate complex,0.21,0.2,0.41000000000000003
+"Gustatory areas, layer 2/3",0.211,0.217,0.428
+Nucleus of reuniens,0.20500000000000002,0.229,0.43400000000000005
+Lateral habenula,0.159,0.161,0.32
+"Gustatory areas, layer 5",0.266,0.279,0.545
+"Accessory olfactory bulb, glomerular layer",0.065,0.079,0.14400000000000002
+Rhomboid nucleus,0.035,0.05,0.085
+pyramid,0.29,0.293,0.583
+Lateral hypothalamic area,1.077,1.075,2.152
+"Accessory olfactory bulb, granular layer",0.08700000000000001,0.11,0.197
+Rostral linear nucleus raphe,0.011,0.04,0.051000000000000004
+pyramidal decussation,0.017,0.043000000000000003,0.060000000000000005
+"Primary somatosensory area, barrel field, layer 2/3",0.738,0.765,1.5030000000000001
+Medial vestibular nucleus,0.873,0.9470000000000001,1.82
+Linear nucleus of the medulla,0.029,0.036000000000000004,0.065
+"Accessory olfactory bulb, mitral layer",0.131,0.149,0.28
+Area postrema,0.019,0.035,0.054000000000000006
+Lateral vestibular nucleus,0.128,0.14400000000000002,0.272
+Lateral mammillary nucleus,0.036000000000000004,0.037,0.07300000000000001
+"Anterior cingulate area, dorsal part, layer 2/3",0.233,0.241,0.474
+Red nucleus,0.362,0.418,0.78
+Anterior pretectal nucleus,0.624,0.635,1.259
+Superior vestibular nucleus,0.159,0.17400000000000002,0.333
+Lateral posterior nucleus of the thalamus,0.598,0.616,1.214
+Arcuate hypothalamic nucleus,0.147,0.163,0.31
+Spinal vestibular nucleus,0.385,0.385,0.77
+Lateral preoptic area,0.246,0.273,0.519
+sensory root of the trigeminal nerve,0.332,0.356,0.688
+Anterior tegmental nucleus,0.018000000000000002,0.017,0.035
+"Anterolateral visual area, layer 5",0.109,0.09,0.199
+"Temporal association areas, layer 4",0.159,0.148,0.307
+solitary tract,0.006,0.005,0.011
+Nucleus raphe pontis,0.034,0.044,0.078
+"Dorsal auditory area, layer 6b",0.015,0.018000000000000002,0.033
+"Midbrain reticular nucleus, retrorubral area",0.074,0.065,0.139
+"Posterior auditory area, layer 6a",0.039,0.041,0.08
+"Lateral septal nucleus, caudal (caudodorsal) part",0.265,0.321,0.5860000000000001
+"Primary auditory area, layer 2/3",0.251,0.23500000000000001,0.486
+"Dorsal auditory area, layer 5",0.199,0.165,0.364
+Anteroventral nucleus of thalamus,0.2,0.213,0.41300000000000003
+"posteromedial visual area, layer 6a",0.07200000000000001,0.066,0.138
+"Lateral septal nucleus, rostral (rostroventral) part",0.961,0.9540000000000001,1.915
+"Nucleus of the lateral olfactory tract, molecular layer",0.053,0.049,0.10200000000000001
+Reticular nucleus of the thalamus,0.732,0.726,1.458
+Anteroventral preoptic nucleus,0.04,0.047,0.087
+"Lateral septal nucleus, ventral part",0.297,0.298,0.595
+"Nucleus of the lateral olfactory tract, pyramidal layer",0.076,0.075,0.151
+"Posterolateral visual area, layer 2/3",0.095,0.11900000000000001,0.21400000000000002
+Nucleus sagulum,0.046,0.054,0.1
+Anteroventral periventricular nucleus,0.078,0.095,0.173
+"Retrosplenial area, dorsal part, layer 6a",0.389,0.374,0.763
+"Retrosplenial area, lateral agranular part, layer 6b",0.019,0.019,0.038
+Barrington's nucleus,0.004,0.007,0.011
+"Anteromedial visual area, layer 1",0.07200000000000001,0.057,0.129
+Suprachiasmatic nucleus,0.038,0.041,0.079
+Bed nucleus of the anterior commissure,0.004,0.005,0.009000000000000001
+"Orbital area, ventrolateral part, layer 2/3",0.263,0.254,0.517
+"Temporal association areas, layer 5",0.463,0.47200000000000003,0.935
+Bed nucleus of the accessory olfactory tract,0.016,0.012,0.028
+"Anterior cingulate area, ventral part, layer 2/3",0.218,0.20700000000000002,0.42500000000000004
+Magnocellular nucleus,0.168,0.185,0.353
+stria terminalis,0.149,0.14100000000000001,0.29000000000000004
+"Basolateral amygdalar nucleus, anterior part",0.388,0.392,0.78
+"Prelimbic area, layer 2/3",0.176,0.183,0.359
+"Primary visual area, layer 6b",0.082,0.101,0.183
+Magnocellular reticular nucleus,0.276,0.263,0.539
+Septofimbrial nucleus,0.216,0.267,0.483
+"Basolateral amygdalar nucleus, posterior part",0.368,0.37,0.738
+Midbrain,3.204,3.451,6.655
+"Agranular insular area, posterior part, layer 6a",0.214,0.20400000000000001,0.41800000000000004
+Supragenual nucleus,0.007,0.009000000000000001,0.016
+"Primary motor area, Layer 1",0.656,0.657,1.3130000000000002
+Subgeniculate nucleus,0.012,0.013000000000000001,0.025
+Suprageniculate nucleus,0.099,0.092,0.191
+superior cerebelar peduncles,0.20500000000000002,0.20800000000000002,0.41300000000000003
+"Basomedial amygdalar nucleus, anterior part",0.402,0.406,0.808
+"Agranular insular area, dorsal part, layer 2/3",0.451,0.452,0.903
+"Retrosplenial area, dorsal part, layer 6b",0.02,0.022,0.041999999999999996
+Accessory supraoptic group,0.001,0.002,0.003
+Septohippocampal nucleus,0.018000000000000002,0.016,0.034
+"Basomedial amygdalar nucleus, posterior part",0.372,0.357,0.729
+"Perirhinal area, layer 6a",0.026000000000000002,0.038,0.064
+superior colliculus commissure,0.012,0.023,0.035
+Subfornical organ,0.007,0.015,0.022
+Substantia innominata,1.573,1.553,3.126
+"Agranular insular area, posterior part, layer 5",0.383,0.388,0.771
+Subparaventricular zone,0.074,0.048,0.122
+supraoptic commissures,0.015,0.016,0.031
+Subceruleus nucleus,0.014,0.015,0.028999999999999998
+Bed nuclei of the stria terminalis,0.687,0.668,1.355
+Medulla,2.581,2.854,5.4350000000000005
+"Agranular insular area, posterior part, layer 6b",0.012,0.009000000000000001,0.021
+Preparasubthalamic nucleus,0.007,0.01,0.017
+Sublaterodorsal nucleus,0.021,0.022,0.043
+Mediodorsal nucleus of thalamus,0.6910000000000001,0.683,1.374
+"Prelimbic area, layer 5",0.504,0.498,1.002
+Parasubthalamic nucleus,0.079,0.088,0.16699999999999998
+Submedial nucleus of the thalamus,0.133,0.157,0.29000000000000004
+"Perirhinal area, layer 6b",0.007,0.009000000000000001,0.016
+Infracerebellar nucleus,0.029,0.025,0.054000000000000006
+"Substantia nigra, compact part",0.1,0.10300000000000001,0.203
+"Posterolateral visual area, layer 6a",0.042,0.049,0.091
+cuneate fascicle,0.007,0.01,0.017
+"Substantia nigra, reticular part",0.778,0.721,1.499
+Field CA1,5.165,5.13,10.295
+Supraoptic nucleus,0.019,0.026000000000000002,0.045
+"Posterolateral visual area, layer 6b",0.004,0.005,0.009000000000000001
+ventral tegmental decussation,0.018000000000000002,0.032,0.05
+"Anteromedial visual area, layer 4",0.032,0.029,0.061
+Medial amygdalar nucleus,1.074,1.072,2.146
+"Orbital area, lateral part, layer 2/3",0.305,0.3,0.605
+vestibular nerve,0.116,0.115,0.231
+"Subparafascicular nucleus, magnocellular part",0.039,0.033,0.07200000000000001
+"Lateral visual area, layer 1",0.099,0.089,0.188
+"Subparafascicular nucleus, parvicellular part",0.075,0.064,0.139
+Field CA2,0.224,0.223,0.447
+Ectorhinal area/Layer 2/3,0.246,0.23900000000000002,0.485
+medial corticohypothalamic tract,0.005,0.005,0.01
+"Spinal nucleus of the trigeminal, caudal part",0.807,0.806,1.613
+"Retrosplenial area, ventral part, layer 2/3",0.539,0.514,1.053
+"Anteromedial visual area, layer 5",0.116,0.121,0.237
+"Retrosplenial area, dorsal part, layer 2/3",0.482,0.483,0.965
+columns of the fornix,0.139,0.147,0.28600000000000003
+"Spinal nucleus of the trigeminal, interpolar part",0.924,0.915,1.839
+"Orbital area, lateral part, layer 6a",0.23600000000000002,0.23500000000000001,0.47100000000000003
+"Anteromedial visual area, layer 6b",0.014,0.015,0.028999999999999998
+"Retrosplenial area, dorsal part, layer 1",0.399,0.418,0.817
+dorsal hippocampal commissure,0.47000000000000003,0.48,0.95
+"Spinal nucleus of the trigeminal, oral part",0.52,0.535,1.0550000000000002
+"Orbital area, lateral part, layer 1",0.181,0.192,0.373
+ventral hippocampal commissure,0.03,0.04,0.07
+"Primary somatosensory area, upper limb, layer 1",0.245,0.268,0.513
+"Basolateral amygdalar nucleus, ventral part",0.229,0.212,0.441
+Median preoptic nucleus,0.002,0.034,0.036000000000000004
+"Posterior auditory area, layer 6b",0.008,0.008,0.016
+Midbrain trigeminal nucleus,0.004,0.006,0.01
+"Primary somatosensory area, trunk, layer 6b",0.021,0.022,0.043
+Field CA3,3.173,3.082,6.255
+alveus,0.6890000000000001,0.7030000000000001,1.3920000000000001
+"posteromedial visual area, layer 6b",0.016,0.016,0.032
+Subthalamic nucleus,0.10300000000000001,0.106,0.20900000000000002
+Striatum,1.334,1.419,2.753
+"Primary somatosensory area, lower limb, layer 6a",0.312,0.243,0.5549999999999999
+brachium of the inferior colliculus,0.113,0.122,0.235
+Medial habenula,0.159,0.15,0.309
+"Orbital area, medial part, layer 1",0.17,0.267,0.43700000000000006
+"Orbital area, lateral part, layer 6b",0.018000000000000002,0.021,0.03900000000000001
+"posteromedial visual area, layer 4",0.039,0.042,0.081
+Subiculum,1.098,1.045,2.143
+dorsal acoustic stria,0.006,0.004,0.01
+Main olfactory bulb,3.379,3.755,7.134
+"Primary somatosensory area, lower limb, layer 6b",0.023,0.023,0.046
+Cerebellum,0.41400000000000003,0.419,0.833
+Medial preoptic nucleus,0.203,0.211,0.41400000000000003
+"Ventral auditory area, layer 6a",0.138,0.14300000000000002,0.281
+Medial preoptic area,0.269,0.28800000000000003,0.557
+Supramammillary nucleus,0.125,0.14,0.265
+"Entorhinal area, medial part, dorsal zone, layer 1",0.644,0.636,1.28
+"Dorsal auditory area, layer 1",0.07200000000000001,0.094,0.166
+dorsal fornix,0.009000000000000001,0.017,0.026000000000000002
+Medial pretectal area,0.024,0.023,0.047
+Supratrigeminal nucleus,0.132,0.116,0.248
+dorsal limb,0.07100000000000001,0.08,0.15100000000000002
+"Perirhinal area, layer 1",0.117,0.113,0.23
+"Retrosplenial area, ventral part, layer 1",0.322,0.5720000000000001,0.8940000000000001
+"Entorhinal area, medial part, dorsal zone, layer 2",0.671,0.657,1.328
+"Central amygdalar nucleus, capsular part",0.147,0.176,0.32299999999999995
+Thalamus,0.439,0.446,0.885
+"Central amygdalar nucleus, lateral part",0.128,0.135,0.263
+dorsal spinocerebellar tract,0.074,0.079,0.153
+"Infralimbic area, layer 2/3",0.078,0.074,0.152
+"Primary somatosensory area, nose, layer 1",0.209,0.20800000000000002,0.41700000000000004
+"Central amygdalar nucleus, medial part",0.365,0.377,0.742
+Medial septal nucleus,0.11900000000000001,0.281,0.4
+"posteromedial visual area, layer 5",0.136,0.146,0.28200000000000003
+Postpiriform transition area,0.634,0.634,1.268
+"Lateral visual area, layer 4",0.096,0.085,0.181
+Tegmental reticular nucleus,0.36,0.343,0.7030000000000001
+Central lateral nucleus of the thalamus,0.166,0.183,0.349
+Accessory facial motor nucleus,0.002,0.001,0.003
+"Primary somatosensory area, upper limb, layer 4",0.264,0.219,0.483
+external capsule,0.443,0.45,0.893
+Nucleus of the brachium of the inferior colliculus,0.043000000000000003,0.047,0.09
+Triangular nucleus of septum,0.16,0.199,0.359
+"Orbital area, medial part, layer 2/3",0.151,0.15,0.301
+Claustrum,0.272,0.276,0.548
+Nucleus of Darkschewitsch,0.059000000000000004,0.062,0.121
+"Anterior cingulate area, ventral part, layer 1",0.194,0.339,0.533
+"Retrosplenial area, ventral part, layer 6a",0.322,0.308,0.63
+Central linear nucleus raphe,0.038,0.063,0.101
+"Primary visual area, layer 1",0.628,0.628,1.256
+fasciculus retroflexus,0.084,0.082,0.166
+Diagonal band nucleus,0.342,0.402,0.744
+"Taenia tecta, dorsal part",0.34700000000000003,0.391,0.738
+"Ventral auditory area, layer 6b",0.021,0.024,0.045
+Central medial nucleus of the thalamus,0.098,0.156,0.254
+"Dorsal auditory area, layer 2/3",0.15,0.14300000000000002,0.29300000000000004
+"Anterolateral visual area, layer 6a",0.061,0.047,0.108
+fimbria,0.787,0.728,1.5150000000000001
+Nucleus incertus,0.035,0.07100000000000001,0.10600000000000001
+"Taenia tecta, ventral part",0.387,0.374,0.761
+"Orbital area, ventrolateral part, layer 6a",0.113,0.10400000000000001,0.21700000000000003
+Subparafascicular area,0.053,0.089,0.142
+"Retrosplenial area, dorsal part, layer 5",0.615,0.603,1.218
+habenular commissure,0.016,0.023,0.039
+Nucleus of the lateral lemniscus,0.387,0.375,0.762
+"Lateral visual area, layer 5",0.167,0.151,0.318
+Tuberal nucleus,0.259,0.293,0.552
+Cuneiform nucleus,0.256,0.28600000000000003,0.542
+"Orbital area, medial part, layer 5",0.222,0.233,0.455
+Motor nucleus of trigeminal,0.183,0.187,0.37
+"Retrosplenial area, ventral part, layer 6b",0.024,0.026000000000000002,0.05
+"Primary somatosensory area, upper limb, layer 5",0.316,0.34500000000000003,0.661
+Nucleus of the optic tract,0.099,0.106,0.20500000000000002
+Ventral anterior-lateral complex of the thalamus,0.401,0.399,0.8
+"Orbital area, lateral part, layer 5",0.601,0.551,1.1520000000000001
+"Dentate gyrus, granule cell layer",0.8170000000000001,0.8220000000000001,1.6390000000000002
+inferior colliculus commissure,0.009000000000000001,0.011,0.02
+Nucleus of the posterior commissure,0.147,0.153,0.3
+"Gustatory areas, layer 6a",0.20800000000000002,0.224,0.43200000000000005
+"Cortical amygdalar area, anterior part",0.40900000000000003,0.401,0.81
+Nucleus of the trapezoid body,0.08,0.08,0.16
+"Posterior auditory area, layer 2/3",0.07200000000000001,0.07100000000000001,0.14300000000000002
+"Primary motor area, Layer 5",1.585,1.6480000000000001,3.233
+"Anterolateral visual area, layer 6b",0.02,0.015,0.035
+Nucleus of the solitary tract,0.421,0.451,0.872
+Abducens nucleus,0.016,0.019,0.035
+"Primary somatosensory area, nose, layer 4",0.263,0.271,0.534
+"Cortical amygdalar area, posterior part, lateral zone",0.599,0.639,1.238
+"Secondary motor area, layer 1",1.235,1.171,2.406
+"Primary somatosensory area, mouth, layer 2/3",0.749,0.757,1.506
+lateral lemniscus,0.434,0.421,0.855
+Facial motor nucleus,0.46,0.487,0.9470000000000001
+"Gustatory areas, layer 6b",0.026000000000000002,0.022,0.048
+"Cortical amygdalar area, posterior part, medial zone",0.636,0.654,1.29
+"Entorhinal area, medial part, dorsal zone, layer 3",0.514,0.498,1.012
+"lateral olfactory tract, body",0.47800000000000004,0.47800000000000004,0.9560000000000001
+"Frontal pole, layer 2/3",0.113,0.125,0.238
+"Primary somatosensory area, trunk, layer 2/3",0.179,0.20800000000000002,0.387
+"Retrosplenial area, lateral agranular part, layer 1",0.241,0.244,0.485
+Caudoputamen,12.943,13.077,26.02
+mammillary peduncle,0.017,0.017,0.034
+"Agranular insular area, ventral part, layer 6a",0.08700000000000001,0.089,0.176
+"Dorsal auditory area, layer 4",0.067,0.059000000000000004,0.126
+Superior central nucleus raphe,0.274,0.343,0.617
+"Orbital area, ventrolateral part, layer 6b",0.003,0.003,0.006
+mammillotegmental tract,0.029,0.024,0.053000000000000005
+Ventral medial nucleus of the thalamus,0.482,0.504,0.986
+"Retrosplenial area, ventral part, layer 5",0.8160000000000001,0.8260000000000001,1.6420000000000001
+Ventrolateral preoptic nucleus,0.036000000000000004,0.03,0.066
+mammillothalamic tract,0.063,0.065,0.128
+"Perirhinal area, layer 5",0.085,0.084,0.169
+Ventromedial hypothalamic nucleus,0.27,0.275,0.545
+"Agranular insular area, ventral part, layer 2/3",0.28700000000000003,0.295,0.5820000000000001
+"Posterior auditory area, layer 1",0.053,0.052000000000000005,0.10500000000000001
+medial lemniscus,0.34800000000000003,0.327,0.675
+Olfactory areas,1.3900000000000001,1.487,2.8770000000000002
+"Agranular insular area, ventral part, layer 6b",0.001,0.001,0.002
+"Primary somatosensory area, nose, layer 5",0.308,0.28800000000000003,0.5960000000000001
+Cortical subplate,0.186,0.195,0.381
+"Agranular insular area, ventral part, layer 1",0.123,0.117,0.24
+Olivary pretectal nucleus,0.03,0.03,0.06
+"Infralimbic area, layer 1",0.044,0.10200000000000001,0.14600000000000002
+Cuneate nucleus,0.167,0.167,0.334
+Ventral posterolateral nucleus of the thalamus,0.47400000000000003,0.48,0.954
+"Primary visual area, layer 4",0.494,0.506,1.0
+"Ventral posterolateral nucleus of the thalamus, parvicellular part",0.04,0.051000000000000004,0.091
+"Entorhinal area, medial part, dorsal zone, layer 5",0.47800000000000004,0.461,0.9390000000000001
+arbor vitae,3.358,3.416,6.774
+"Temporal association areas, layer 6a",0.258,0.261,0.519
+"Medial mammillary nucleus, median part",0.025,0.053,0.078
+Ventral posteromedial nucleus of the thalamus,0.8200000000000001,0.845,1.665
+"Primary auditory area, layer 1",0.2,0.185,0.385
+"Ventral posteromedial nucleus of the thalamus, parvicellular part",0.11900000000000001,0.139,0.258
+"Entorhinal area, medial part, dorsal zone, layer 6",0.337,0.317,0.654
+cerebellar commissure,0.028,0.033,0.061
+Ventral tegmental area,0.222,0.226,0.448
+"Posterolateral visual area, layer 1",0.096,0.099,0.195
+principal mammillary tract,0.017,0.021,0.038000000000000006
+Olfactory tubercle,1.982,1.993,3.975
+"Ventral auditory area, layer 2/3",0.192,0.209,0.401
+Ventral tegmental nucleus,0.014,0.015,0.028999999999999998
+"Posterior auditory area, layer 4",0.044,0.038,0.08199999999999999
+Nucleus x,0.027,0.027,0.054
+"Secondary motor area, layer 5",2.351,2.101,4.452
+Pons,1.722,1.951,3.673
+"Anterior cingulate area, ventral part, layer 5",0.559,0.546,1.105
+Hypoglossal nucleus,0.131,0.14100000000000001,0.272
+"Retrosplenial area, lateral agranular part, layer 5",0.36,0.359,0.719
+"Primary visual area, layer 5",0.756,0.773,1.529
+Posterior amygdalar nucleus,0.47900000000000004,0.488,0.9670000000000001
+Nucleus y,0.011,0.013000000000000001,0.024
+"Agranular insular area, dorsal part, layer 6a",0.397,0.393,0.79
+corticospinal tract,0.044,0.049,0.093
+"Temporal association areas, layer 6b",0.042,0.032,0.07400000000000001
+Piriform-amygdalar area,0.607,0.648,1.255
+"Posterior auditory area, layer 5",0.093,0.095,0.188
+spinal tract of the trigeminal nerve,0.846,0.838,1.684
+Periaqueductal gray,1.962,2.165,4.127
+Zona incerta,0.782,0.726,1.508
+facial nerve,0.039,0.039,0.078
+"Agranular insular area, ventral part, layer 5",0.36,0.359,0.719
+stria medullaris,0.14100000000000001,0.134,0.275
+Pallidum,0.5760000000000001,0.5680000000000001,1.1440000000000001
+Fields of Forel,0.132,0.109,0.241
+"posteromedial visual area, layer 1",0.088,0.098,0.186
+"Supplemental somatosensory area, layer 2/3",1.061,1.029,2.09
+"Anterior cingulate area, ventral part, 6a",0.198,0.176,0.374
+"Inferior colliculus, central nucleus",0.535,0.5730000000000001,1.108
+superior cerebellar peduncle decussation,0.013000000000000001,0.03,0.043
+Dorsal peduncular area,0.234,0.267,0.501
+"Primary auditory area, layer 4",0.125,0.114,0.239
+"Anterior cingulate area, ventral part, 6b",0.035,0.029,0.064
+"Inferior colliculus, dorsal nucleus",0.664,0.652,1.316
+"Primary visual area, layer 2/3",0.975,0.981,1.956
+"Infralimbic area, layer 5",0.14400000000000002,0.135,0.279
+"Inferior colliculus, external nucleus",1.016,1.004,2.02
+Dorsomedial nucleus of the hypothalamus,0.21,0.178,0.388
+"Agranular insular area, dorsal part, layer 6b",0.026000000000000002,0.026000000000000002,0.052000000000000005
+oculomotor nerve,0.004,0.005,0.009000000000000001
+"Superior colliculus, zonal layer",0.135,0.169,0.30400000000000005
+Ectorhinal area/Layer 1,0.132,0.151,0.28300000000000003
+"Primary somatosensory area, nose, layer 2/3",0.312,0.329,0.641
+Dorsal motor nucleus of the vagus nerve,0.08600000000000001,0.09,0.176
+trapezoid body,0.157,0.184,0.34099999999999997
+"Superior colliculus, superficial gray layer",0.586,0.595,1.181
+Parasubiculum,0.48,0.458,0.938
+"Primary motor area, Layer 6a",1.333,1.4020000000000001,2.7350000000000003
+Dentate nucleus,0.183,0.156,0.33899999999999997
+"Primary auditory area, layer 5",0.34800000000000003,0.335,0.683
+optic nerve,0.034,0.04,0.07400000000000001
+"Visceral area, layer 6b",0.022,0.02,0.041999999999999996
+uncinate fascicle,0.04,0.039,0.079
+"Superior colliculus, optic layer",0.34500000000000003,0.31,0.655
+Parvicellular reticular nucleus,1.171,1.205,2.3760000000000003
+"Primary somatosensory area, upper limb, layer 2/3",0.511,0.453,0.964
+"Visceral area, layer 6a",0.278,0.266,0.544
+Parasolitary nucleus,0.017,0.014,0.031
+"Supplemental somatosensory area, layer 6a",0.998,0.992,1.99
+rubrospinal tract,0.302,0.29,0.592
+ventral spinocerebellar tract,0.159,0.161,0.32
+Parabrachial nucleus,0.453,0.486,0.9390000000000001
+"Posterolateral visual area, layer 4",0.016,0.019,0.035
+Dorsal nucleus raphe,0.026000000000000002,0.115,0.14100000000000001
+"Supplemental somatosensory area, layer 1",0.666,0.628,1.294
+Parabigeminal nucleus,0.024,0.024,0.048
+"Primary somatosensory area, mouth, layer 1",0.382,0.387,0.769
+Dorsal tegmental nucleus,0.051000000000000004,0.06,0.111
+"Primary motor area, Layer 6b",0.096,0.099,0.195
+amygdalar capsule,0.089,0.093,0.182
+"Perirhinal area, layer 2/3",0.158,0.171,0.329
+"Primary somatosensory area, nose, layer 6a",0.43,0.39,0.8200000000000001
+"Supplemental somatosensory area, layer 6b",0.105,0.092,0.197
+"Visceral area, layer 1",0.168,0.151,0.319
+Pontine central gray,0.258,0.271,0.529
+"anterior commissure, olfactory limb",0.371,0.422,0.7929999999999999
+"Posterolateral visual area, layer 5",0.10400000000000001,0.13,0.234
+External cuneate nucleus,0.10400000000000001,0.098,0.202
+"Anterolateral visual area, layer 2/3",0.096,0.098,0.194
+"Retrosplenial area, lateral agranular part, layer 6a",0.20800000000000002,0.202,0.41000000000000003
+Paracentral nucleus,0.115,0.112,0.227
+"anterior commissure, temporal limb",0.121,0.113,0.23399999999999999
+"Orbital area, medial part, layer 6a",0.09,0.08600000000000001,0.176
+trochlear nerve,0.004,0.005,0.009000000000000001
+Lingula (I),0.055,0.076,0.131
+Posterodorsal preoptic nucleus,0.005,0.004,0.009000000000000001
+brachium of the superior colliculus,0.088,0.097,0.185
+"Anterior cingulate area, dorsal part, layer 6a",0.332,0.385,0.7170000000000001
+cerebal peduncle,0.488,0.492,0.98
+"Anterior cingulate area, dorsal part, layer 6b",0.011,0.012,0.023
+"Primary somatosensory area, nose, layer 6b",0.026000000000000002,0.026000000000000002,0.052000000000000005
+Parafascicular nucleus,0.23900000000000002,0.255,0.494
+Pontine gray,0.485,0.496,0.981
+"Anterior cingulate area, dorsal part, layer 1",0.214,0.367,0.581
+Declive (VI),1.6,1.605,3.205
+"Nucleus ambiguus, dorsal division",0.014,0.014,0.028
+cingulum bundle,0.645,0.613,1.258
+"Primary motor area, Layer 2/3",1.9180000000000001,1.83,3.748
+Folium-tuber vermis (VII),0.549,0.518,1.0670000000000002
+"Primary somatosensory area, upper limb, layer 6a",0.507,0.497,1.004
+Posterior hypothalamic nucleus,0.322,0.35100000000000003,0.673
+vomeronasal nerve,0.008,0.008,0.016
+"Primary somatosensory area, mouth, layer 4",0.452,0.485,0.937
+Pyramus (VIII),0.582,0.581,1.1629999999999998
+"Endopiriform nucleus, dorsal part",0.925,0.901,1.826
+"Primary auditory area, layer 6a",0.147,0.139,0.28600000000000003
+"Lateral reticular nucleus, magnocellular part",0.24,0.263,0.503
+"corpus callosum, anterior forceps",0.437,0.406,0.843
+Uvula (IX),1.054,1.112,2.1660000000000004
+"Ventral auditory area, layer 1",0.14100000000000001,0.134,0.275
+Piriform area,5.931,5.878,11.809000000000001
+"Secondary motor area, layer 2/3",1.923,1.872,3.795
+"Lateral reticular nucleus, parvicellular part",0.029,0.034,0.063
+"corpus callosum, extreme capsule",0.054,0.048,0.10200000000000001
+"Retrosplenial area, lateral agranular part, layer 2/3",0.303,0.325,0.628
+"Endopiriform nucleus, ventral part",0.501,0.491,0.992
+Nodulus (X),0.731,0.797,1.528
+"Orbital area, ventrolateral part, layer 1",0.2,0.197,0.397
+"Paragigantocellular reticular nucleus, dorsal part",0.117,0.121,0.238
+"corpus callosum, posterior forceps",0.668,0.66,1.328
+"Lateral visual area, layer 2/3",0.158,0.149,0.307
+"Primary somatosensory area, mouth, layer 5",0.5740000000000001,0.614,1.1880000000000002
+Lobule II,0.612,0.717,1.329
+Ectorhinal area/Layer 6a,0.2,0.185,0.385
+"Paragigantocellular reticular nucleus, lateral part",0.383,0.361,0.744
+Dorsal premammillary nucleus,0.064,0.074,0.138
+"Primary somatosensory area, barrel field, layer 1",0.4,0.40700000000000003,0.807
+Fasciola cinerea,0.028,0.029,0.057
+Lobule III,1.281,1.456,2.737
+"corpus callosum, splenium",0.34400000000000003,0.363,0.7070000000000001
+Ectorhinal area/Layer 5,0.279,0.249,0.528
+Fastigial nucleus,0.248,0.275,0.523
+"Ventral auditory area, layer 4",0.108,0.107,0.215
+"Agranular insular area, dorsal part, layer 1",0.233,0.228,0.461
+root,1.731,1.877,3.608
+Fundus of striatum,0.242,0.221,0.46299999999999997
+Ventral premammillary nucleus,0.11,0.112,0.222
+"Primary auditory area, layer 6b",0.018000000000000002,0.019,0.037000000000000005
+"Primary somatosensory area, trunk, layer 1",0.091,0.101,0.192
+Simple lobule,2.824,2.695,5.519
+fiber tracts,0.807,0.802,1.609
+"Visceral area, layer 4",0.097,0.094,0.191
+"Anterior cingulate area, dorsal part, layer 5",0.5630000000000001,0.582,1.145
+olfactory nerve layer of main olfactory bulb,0.433,0.46,0.893
+Posterior complex of the thalamus,0.631,0.622,1.2530000000000001
+"Secondary motor area, layer 6a",1.038,1.236,2.274
+"Globus pallidus, external segment",0.791,0.8,1.5910000000000002
+"Ventral auditory area, layer 5",0.303,0.301,0.604
+Paramedian lobule,2.482,2.471,4.953
+"Primary somatosensory area, upper limb, layer 6b",0.042,0.043000000000000003,0.085
+Posterior limiting nucleus of the thalamus,0.097,0.096,0.193
+"Primary somatosensory area, lower limb, layer 1",0.17300000000000001,0.14,0.31300000000000006
+"Globus pallidus, internal segment",0.20400000000000001,0.194,0.398
+Copula pyramidis,1.216,1.173,2.3890000000000002
+"Supplemental somatosensory area, layer 4",0.5740000000000001,0.5690000000000001,1.1430000000000002
+Postsubiculum,0.515,0.52,1.0350000000000001
+"Primary somatosensory area, barrel field, layer 6a",0.636,0.633,1.2690000000000001
+Gracile nucleus,0.037,0.042,0.079
+Paraflocculus,2.949,2.866,5.8149999999999995
+crossed tectospinal pathway,0.248,0.255,0.503
+Peripeduncular nucleus,0.036000000000000004,0.028,0.064
+Ectorhinal area/Layer 6b,0.033,0.024,0.057
+"Anteromedial visual area, layer 6a",0.057,0.067,0.124
+"Primary somatosensory area, barrel field, layer 4",0.683,0.652,1.335
+Gigantocellular reticular nucleus,1.234,1.352,2.5860000000000003
+Flocculus,0.671,0.676,1.347
+Pedunculopontine nucleus,0.482,0.405,0.887
+"Infralimbic area, layer 6a",0.108,0.093,0.201
+Crus 1,2.846,2.797,5.643000000000001
+"Visceral area, layer 5",0.374,0.34800000000000003,0.722
+Posterior pretectal nucleus,0.07100000000000001,0.073,0.14400000000000002
+"Primary somatosensory area, barrel field, layer 6b",0.06,0.075,0.135
+Crus 2,2.516,2.605,5.121
+"Anteromedial visual area, layer 2/3",0.115,0.094,0.20900000000000002
+Parapyramidal nucleus,0.053,0.049,0.10200000000000001
+"Primary somatosensory area, barrel field, layer 5",0.628,0.59,1.218
+"Medial geniculate complex, dorsal part",0.082,0.083,0.165
+"Anterolateral visual area, layer 1",0.053,0.067,0.12
+Perireunensis nucleus,0.078,0.082,0.16
+"Medial geniculate complex, ventral part",0.128,0.133,0.261
+"Infralimbic area, layer 6b",0.004,0.003,0.007
+Presubiculum,0.465,0.463,0.928
+"Secondary motor area, layer 6b",0.036000000000000004,0.039,0.07500000000000001
+"Primary somatosensory area, trunk, layer 4",0.067,0.074,0.14100000000000001
+"Medial geniculate complex, medial part",0.11,0.11900000000000001,0.229
+Hippocampal formation,0.196,0.219,0.41500000000000004
+"Supplemental somatosensory area, layer 5",1.049,1.055,2.104
+Lobules IV-V,3.306,3.3890000000000002,6.695
+external medullary lamina of the thalamus,0.054,0.054,0.108
+"Pontine reticular nucleus, caudal part",1.2,1.193,2.393
+"Primary somatosensory area, lower limb, layer 4",0.11900000000000001,0.14100000000000001,0.26
+"Anteromedial nucleus, dorsal part",0.13,0.145,0.275
+Hypothalamus,1.3920000000000001,1.413,2.805
+"Medullary reticular nucleus, dorsal part",0.527,0.537,1.064
+"Agranular insular area, dorsal part, layer 5",0.755,0.747,1.502
+"Primary somatosensory area, mouth, layer 6a",0.891,0.805,1.6960000000000002
+"Anteromedial nucleus, ventral part",0.096,0.081,0.177
+Intercalated amygdalar nucleus,0.097,0.088,0.185
+"Visceral area, layer 2/3",0.278,0.264,0.542
+"Medullary reticular nucleus, ventral part",0.483,0.46900000000000003,0.952
+genu of corpus callosum,0.41300000000000003,0.386,0.799
+Parastrial nucleus,0.061,0.043000000000000003,0.10400000000000001
+"Primary somatosensory area, trunk, layer 5",0.179,0.17300000000000001,0.352
+Interanterodorsal nucleus of the thalamus,0.06,0.066,0.126
+"Anterolateral visual area, layer 4",0.059000000000000004,0.048,0.10700000000000001
+genu of the facial nerve,0.015,0.018000000000000002,0.033
+Interanteromedial nucleus of the thalamus,0.026000000000000002,0.033,0.059000000000000004
+"Entorhinal area, lateral part, layer 1",0.487,0.49,0.977
+inferior cerebellar peduncle,0.374,0.369,0.743
+"Orbital area, ventrolateral part, layer 5",0.319,0.31,0.629
+"Tuberomammillary nucleus, dorsal part",0.021,0.024,0.045
+"Temporal association areas, layer 2/3",0.367,0.34900000000000003,0.716
+"Primary somatosensory area, lower limb, layer 5",0.226,0.276,0.502
+"Nucleus of the lateral olfactory tract, layer 3",0.036000000000000004,0.031,0.067
+Median eminence,0.033,0.055,0.088
+"Dentate gyrus, molecular layer",2.239,2.1350000000000002,4.3740000000000006
+"Dentate gyrus, polymorph layer",0.32,0.311,0.631
+"Primary somatosensory area, unassigned, layer 1",0.085,0.066,0.15100000000000002
+"Primary somatosensory area, unassigned, layer 2/3",0.168,0.131,0.29900000000000004
+"Primary somatosensory area, unassigned, layer 4",0.07,0.10300000000000001,0.17300000000000001
+"Primary somatosensory area, unassigned, layer 5",0.124,0.152,0.276
+"Primary somatosensory area, unassigned, layer 6a",0.166,0.147,0.313
+"Primary somatosensory area, unassigned, layer 6b",0.015,0.015,0.03
+"Anterior area, layer 1",0.107,0.113,0.22
+"Anterior area, layer 2/3",0.215,0.213,0.428
+"Anterior area, layer 4",0.085,0.081,0.166
+"Anterior area, layer 5",0.213,0.17500000000000002,0.388
+"Anterior area, layer 6a",0.11,0.108,0.218
+"Anterior area, layer 6b",0.022,0.026000000000000002,0.048
+"Laterointermediate area, layer 1",0.037,0.035,0.07200000000000001
+"Laterointermediate area, layer 2/3",0.064,0.057,0.121
+"Laterointermediate area, layer 4",0.028,0.03,0.057999999999999996
+"Laterointermediate area, layer 5",0.078,0.073,0.151
+"Laterointermediate area, layer 6a",0.04,0.035,0.07500000000000001
+"Laterointermediate area, layer 6b",0.009000000000000001,0.008,0.017
+"Rostrolateral area, layer 1",0.088,0.08600000000000001,0.174
+"Rostrolateral area, layer 2/3",0.14200000000000002,0.132,0.274
+"Rostrolateral area, layer 4",0.07,0.07200000000000001,0.14200000000000002
+"Rostrolateral area, layer 5",0.113,0.123,0.236
+"Rostrolateral area, layer 6a",0.075,0.078,0.153
+"Rostrolateral area, layer 6b",0.009000000000000001,0.018000000000000002,0.027000000000000003
+"Postrhinal area, layer 1",0.115,0.121,0.236
+"Postrhinal area, layer 2/3",0.188,0.17500000000000002,0.363
+"Postrhinal area, layer 4",0.031,0.032,0.063
+"Postrhinal area, layer 5",0.181,0.191,0.372
+"Postrhinal area, layer 6a",0.08700000000000001,0.093,0.18
+"Postrhinal area, layer 6b",0.018000000000000002,0.018000000000000002,0.036000000000000004
+Prosubiculum,0.6,0.595,1.1949999999999998
+Area prostriata,0.17200000000000001,0.185,0.357
+supra-callosal cerebral white matter,0.561,0.514,1.0750000000000002
+"corpus callosum, body",1.5090000000000001,1.362,2.8710000000000004
+optic radiation,0.927,0.897,1.824
+auditory radiation,0.218,0.221,0.439
+commissural branch of stria terminalis,0.017,0.019,0.036000000000000004
+"Dorsal part of the lateral geniculate complex, shell",0.101,0.1,0.201
+"Dorsal part of the lateral geniculate complex, core",0.211,0.20600000000000002,0.41700000000000004
+"Dorsal part of the lateral geniculate complex, ipsilateral zone",0.043000000000000003,0.042,0.085
+"Frontal pole, layer 5",0.183,0.159,0.34199999999999997
+"Frontal pole, layer 6a",0.061,0.056,0.11699999999999999
+"Frontal pole, layer 6b",0.001,0.001,0.002
+"Orbital area, medial part, layer 6b",0.003,0.002,0.005
+Retroparafascicular nucleus,0.03,0.029,0.059
+Medial accesory oculomotor nucleus,0.01,0.01,0.02
+Peritrigeminal zone,0.177,0.149,0.32599999999999996
+Accessory trigeminal nucleus,0.007,0.009000000000000001,0.016
+Parvicellular motor 5 nucleus,0.033,0.034,0.067
+Intertrigeminal nucleus,0.023,0.029,0.052000000000000005
+Ethmoid nucleus of the thalamus,0.129,0.11900000000000001,0.248
+Xiphoid thalamic nucleus,0.006,0.066,0.07200000000000001
+Posterior intralaminar thalamic nucleus,0.099,0.084,0.183
+Posterior triangular thalamic nucleus,0.126,0.14,0.266
+Intermediate geniculate nucleus,0.011,0.01,0.020999999999999998
+Ventromedial preoptic nucleus,0.022,0.022,0.044
+Perifornical nucleus,0.115,0.101,0.21600000000000003
+Hippocampo-amygdalar transition area,0.21,0.195,0.405
+Paratrigeminal nucleus,0.053,0.037,0.09
+Vestibulocerebellar nucleus,0.041,0.046,0.087
+Subcommissural organ,0.002,0.01,0.012
+Posterodorsal tegmental nucleus,0.019,0.017,0.036000000000000004
+"Medial mammillary nucleus, lateral part",0.11900000000000001,0.112,0.231
+"Medial mammillary nucleus, medial part",0.081,0.078,0.159
+"Medial mammillary nucleus, posterior part",0.012,0.017,0.029
+"Medial mammillary nucleus, dorsal part",0.034,0.048,0.082
+Paratrochlear nucleus,0.011,0.009000000000000001,0.02
+Paranigral nucleus,0.012,0.011,0.023
+"Interpeduncular nucleus, rostral",0.029,0.051000000000000004,0.08
+"Interpeduncular nucleus, caudal",0.021,0.044,0.065
+"Interpeduncular nucleus, apical",0.005,0.015,0.02
+"Interpeduncular nucleus, lateral",0.026000000000000002,0.03,0.056
+"Interpeduncular nucleus, intermediate",0.021,0.021,0.042
+"Interpeduncular nucleus, dorsomedial",0.011,0.011,0.022
+"Interpeduncular nucleus, dorsolateral",0.018000000000000002,0.016,0.034
+"Interpeduncular nucleus, rostrolateral",0.009000000000000001,0.009000000000000001,0.018000000000000002
+Supraoculomotor periaqueductal gray,0.018000000000000002,0.016,0.034
diff --git a/tests/data/registration_output/Windows/boundaries.tiff b/tests/data/registration_output/Windows/boundaries.tiff
new file mode 100644
index 00000000..d11336e9
Binary files /dev/null and b/tests/data/registration_output/Windows/boundaries.tiff differ
diff --git a/tests/data/registration_output/Windows/deformation_field_0.tiff b/tests/data/registration_output/Windows/deformation_field_0.tiff
new file mode 100644
index 00000000..3cf9586a
Binary files /dev/null and b/tests/data/registration_output/Windows/deformation_field_0.tiff differ
diff --git a/tests/data/registration_output/Windows/deformation_field_1.tiff b/tests/data/registration_output/Windows/deformation_field_1.tiff
new file mode 100644
index 00000000..0b461762
Binary files /dev/null and b/tests/data/registration_output/Windows/deformation_field_1.tiff differ
diff --git a/tests/data/registration_output/Windows/deformation_field_2.tiff b/tests/data/registration_output/Windows/deformation_field_2.tiff
new file mode 100644
index 00000000..0abf8b09
Binary files /dev/null and b/tests/data/registration_output/Windows/deformation_field_2.tiff differ
diff --git a/tests/data/registration_output/Windows/downsampled.tiff b/tests/data/registration_output/Windows/downsampled.tiff
new file mode 100644
index 00000000..99486807
Binary files /dev/null and b/tests/data/registration_output/Windows/downsampled.tiff differ
diff --git a/tests/data/registration_output/Windows/downsampled_channel_0.tiff b/tests/data/registration_output/Windows/downsampled_channel_0.tiff
new file mode 100644
index 00000000..99486807
Binary files /dev/null and b/tests/data/registration_output/Windows/downsampled_channel_0.tiff differ
diff --git a/tests/data/registration_output/Windows/downsampled_standard.tiff b/tests/data/registration_output/Windows/downsampled_standard.tiff
new file mode 100644
index 00000000..3a0f3566
Binary files /dev/null and b/tests/data/registration_output/Windows/downsampled_standard.tiff differ
diff --git a/tests/data/registration_output/Windows/downsampled_standard_channel_0.tiff b/tests/data/registration_output/Windows/downsampled_standard_channel_0.tiff
new file mode 100644
index 00000000..3a0f3566
Binary files /dev/null and b/tests/data/registration_output/Windows/downsampled_standard_channel_0.tiff differ
diff --git a/tests/data/registration_output/Windows/registered_atlas.tiff b/tests/data/registration_output/Windows/registered_atlas.tiff
new file mode 100644
index 00000000..9dd642a0
Binary files /dev/null and b/tests/data/registration_output/Windows/registered_atlas.tiff differ
diff --git a/tests/data/registration_output/Windows/registered_hemispheres.tiff b/tests/data/registration_output/Windows/registered_hemispheres.tiff
new file mode 100644
index 00000000..dac21ee1
Binary files /dev/null and b/tests/data/registration_output/Windows/registered_hemispheres.tiff differ
diff --git a/tests/data/registration_output/Windows/volumes.csv b/tests/data/registration_output/Windows/volumes.csv
new file mode 100644
index 00000000..e69a6061
--- /dev/null
+++ b/tests/data/registration_output/Windows/volumes.csv
@@ -0,0 +1,664 @@
+structure_name,left_volume_mm3,right_volume_mm3,total_volume_mm3
+"Tuberomammillary nucleus, ventral part",0.059000000000000004,0.058,0.117
+"Primary somatosensory area, mouth, layer 6b",0.06,0.059000000000000004,0.119
+internal capsule,1.032,0.996,2.028
+Principal sensory nucleus of the trigeminal,0.578,0.537,1.115
+"Primary somatosensory area, trunk, layer 6a",0.117,0.10200000000000001,0.21900000000000003
+"Superior colliculus, motor related, intermediate gray layer",0.9560000000000001,1.008,1.964
+Interfascicular nucleus raphe,0.031,0.053,0.08399999999999999
+Parataenial nucleus,0.126,0.123,0.249
+"Superior colliculus, motor related, intermediate white layer",1.0150000000000001,1.049,2.064
+Induseum griseum,0.037,0.077,0.11399999999999999
+"Entorhinal area, lateral part, layer 2",0.8160000000000001,0.784,1.6
+Anterior amygdalar area,0.243,0.302,0.5449999999999999
+"Superior colliculus, motor related, deep gray layer",0.58,0.5740000000000001,1.154
+Intergeniculate leaflet of the lateral geniculate complex,0.037,0.03,0.067
+"Entorhinal area, lateral part, layer 6a",0.545,0.51,1.0550000000000002
+"Primary visual area, layer 6a",0.542,0.514,1.056
+Oculomotor nucleus,0.011,0.015,0.026
+"Gustatory areas, layer 1",0.101,0.089,0.19
+Paraventricular hypothalamic nucleus,0.095,0.10200000000000001,0.197
+"posteromedial visual area, layer 2/3",0.14200000000000002,0.147,0.28900000000000003
+"Superior colliculus, motor related, deep white layer",0.149,0.15,0.299
+Precommissural nucleus,0.1,0.096,0.196
+"Entorhinal area, lateral part, layer 3",0.682,0.636,1.318
+medial forebrain bundle,0.032,0.024,0.056
+Nucleus accumbens,2.248,2.188,4.436
+Medial terminal nucleus of the accessory optic tract,0.028,0.023,0.051000000000000004
+Intermediodorsal nucleus of the thalamus,0.045,0.138,0.183
+medial longitudinal fascicle,0.045,0.044,0.089
+"Paraventricular hypothalamic nucleus, descending division",0.07200000000000001,0.059000000000000004,0.131
+Anterodorsal nucleus,0.079,0.07200000000000001,0.15100000000000002
+Lateral terminal nucleus of the accessory optic tract,0.01,0.013000000000000001,0.023
+Interstitial nucleus of Cajal,0.041,0.039,0.08
+"Frontal pole, layer 1",0.115,0.10200000000000001,0.21700000000000003
+Anterodorsal preoptic nucleus,0.046,0.039,0.08499999999999999
+"Lateral visual area, layer 6a",0.107,0.094,0.201
+Dorsal terminal nucleus of the accessory optic tract,0.006,0.007,0.013000000000000001
+middle cerebellar peduncle,0.518,0.548,1.066
+lateral ventricle,1.016,1.038,2.0540000000000003
+Inferior olivary complex,0.253,0.234,0.487
+"Prelimbic area, layer 6a",0.232,0.20800000000000002,0.44000000000000006
+Anterior hypothalamic nucleus,0.382,0.341,0.7230000000000001
+Interposed nucleus,0.417,0.41100000000000003,0.8280000000000001
+motor root of the trigeminal nerve,0.027,0.035,0.062
+Dorsal cochlear nucleus,0.312,0.32,0.632
+"Temporal association areas, layer 1",0.263,0.23800000000000002,0.501
+subependymal zone,0.059000000000000004,0.06,0.119
+Interpeduncular nucleus,0.004,0.007,0.011
+Ventral cochlear nucleus,0.512,0.518,1.03
+nigrostriatal tract,0.053,0.06,0.11299999999999999
+"Superior olivary complex, medial part",0.10400000000000001,0.098,0.202
+Inferior salivatory nucleus,0.005,0.005,0.01
+choroid plexus,0.627,0.746,1.373
+"Primary somatosensory area, lower limb, layer 2/3",0.325,0.278,0.603
+"Superior olivary complex, lateral part",0.182,0.17300000000000001,0.355
+Trochlear nucleus,0.003,0.005,0.008
+optic chiasm,0.145,0.167,0.312
+"Periventricular hypothalamic nucleus, intermediate part",0.027,0.124,0.151
+"Agranular insular area, posterior part, layer 1",0.221,0.2,0.42100000000000004
+"Lateral visual area, layer 6b",0.018000000000000002,0.02,0.038000000000000006
+"Superior olivary complex, periolivary region",0.194,0.188,0.382
+Koelliker-Fuse subnucleus,0.096,0.092,0.188
+optic tract,0.34700000000000003,0.367,0.714
+"Periventricular hypothalamic nucleus, posterior part",0.038,0.058,0.096
+Midbrain reticular nucleus,2.6350000000000002,2.509,5.144
+third ventricle,0.23800000000000002,0.768,1.006
+Lateral amygdalar nucleus,0.424,0.402,0.8260000000000001
+"Prelimbic area, layer 6b",0.012,0.007,0.019
+"Periventricular hypothalamic nucleus, preoptic part",0.02,0.1,0.12000000000000001
+Intermediate reticular nucleus,1.416,1.355,2.771
+"Entorhinal area, lateral part, layer 5",0.723,0.6960000000000001,1.419
+cerebral aqueduct,0.146,0.22,0.366
+"Nucleus ambiguus, ventral division",0.007,0.008,0.015
+fourth ventricle,0.226,0.273,0.499
+Pontine reticular nucleus,1.189,1.167,2.356
+Locus ceruleus,0.009000000000000001,0.005,0.014000000000000002
+"Gustatory areas, layer 4",0.07200000000000001,0.056,0.128
+Paraventricular nucleus of the thalamus,0.176,0.28500000000000003,0.461
+lateral recess,0.233,0.229,0.462
+Lateral dorsal nucleus of thalamus,0.501,0.521,1.022
+"Dorsal auditory area, layer 6a",0.10400000000000001,0.084,0.188
+posterior commissure,0.016,0.031,0.047
+Anterior olfactory nucleus,2.315,2.447,4.7620000000000005
+Laterodorsal tegmental nucleus,0.097,0.079,0.176
+"Agranular insular area, posterior part, layer 2/3",0.405,0.376,0.781
+Nucleus prepositus,0.113,0.101,0.21400000000000002
+"Prelimbic area, layer 1",0.194,0.317,0.511
+Retrochiasmatic area,0.067,0.075,0.14200000000000002
+Nucleus of Roller,0.017,0.02,0.037000000000000005
+Ventral part of the lateral geniculate complex,0.203,0.193,0.396
+"Gustatory areas, layer 2/3",0.20600000000000002,0.177,0.383
+Nucleus of reuniens,0.192,0.232,0.42400000000000004
+Lateral habenula,0.147,0.177,0.32399999999999995
+"Gustatory areas, layer 5",0.27,0.244,0.514
+"Accessory olfactory bulb, glomerular layer",0.075,0.073,0.148
+Rhomboid nucleus,0.028,0.051000000000000004,0.079
+pyramid,0.289,0.28400000000000003,0.573
+Lateral hypothalamic area,1.102,1.068,2.17
+"Accessory olfactory bulb, granular layer",0.08700000000000001,0.112,0.199
+Rostral linear nucleus raphe,0.017,0.055,0.07200000000000001
+pyramidal decussation,0.017,0.045,0.062
+"Primary somatosensory area, barrel field, layer 2/3",0.724,0.675,1.399
+Medial vestibular nucleus,0.9,0.913,1.8130000000000002
+Linear nucleus of the medulla,0.03,0.039,0.069
+"Accessory olfactory bulb, mitral layer",0.124,0.132,0.256
+Area postrema,0.016,0.034,0.05
+Lateral vestibular nucleus,0.14,0.138,0.278
+Lateral mammillary nucleus,0.038,0.038,0.076
+"Anterior cingulate area, dorsal part, layer 2/3",0.222,0.218,0.44
+Red nucleus,0.371,0.436,0.8069999999999999
+Anterior pretectal nucleus,0.638,0.624,1.262
+Superior vestibular nucleus,0.17,0.166,0.336
+Lateral posterior nucleus of the thalamus,0.588,0.618,1.206
+Arcuate hypothalamic nucleus,0.147,0.152,0.299
+Spinal vestibular nucleus,0.393,0.382,0.775
+Lateral preoptic area,0.226,0.258,0.484
+sensory root of the trigeminal nerve,0.317,0.34600000000000003,0.663
+Anterior tegmental nucleus,0.016,0.022,0.038
+"Anterolateral visual area, layer 5",0.1,0.093,0.193
+"Temporal association areas, layer 4",0.159,0.137,0.29600000000000004
+solitary tract,0.005,0.006,0.011
+Nucleus raphe pontis,0.034,0.046,0.08
+"Dorsal auditory area, layer 6b",0.016,0.016,0.032
+"Midbrain reticular nucleus, retrorubral area",0.081,0.053,0.134
+"Posterior auditory area, layer 6a",0.043000000000000003,0.038,0.081
+"Lateral septal nucleus, caudal (caudodorsal) part",0.268,0.28800000000000003,0.556
+"Primary auditory area, layer 2/3",0.23800000000000002,0.221,0.459
+"Dorsal auditory area, layer 5",0.166,0.149,0.315
+Anteroventral nucleus of thalamus,0.193,0.222,0.41500000000000004
+"posteromedial visual area, layer 6a",0.07200000000000001,0.07200000000000001,0.14400000000000002
+"Lateral septal nucleus, rostral (rostroventral) part",0.896,0.909,1.8050000000000002
+"Nucleus of the lateral olfactory tract, molecular layer",0.05,0.05,0.1
+Reticular nucleus of the thalamus,0.706,0.6970000000000001,1.403
+Anteroventral preoptic nucleus,0.042,0.048,0.09
+"Lateral septal nucleus, ventral part",0.289,0.301,0.59
+"Nucleus of the lateral olfactory tract, pyramidal layer",0.074,0.075,0.149
+"Posterolateral visual area, layer 2/3",0.092,0.096,0.188
+Nucleus sagulum,0.04,0.055,0.095
+Anteroventral periventricular nucleus,0.092,0.10300000000000001,0.195
+"Retrosplenial area, dorsal part, layer 6a",0.384,0.376,0.76
+"Retrosplenial area, lateral agranular part, layer 6b",0.019,0.017,0.036000000000000004
+Barrington's nucleus,0.008,0.007,0.015
+"Anteromedial visual area, layer 1",0.077,0.068,0.14500000000000002
+Suprachiasmatic nucleus,0.031,0.034,0.065
+Bed nucleus of the anterior commissure,0.004,0.002,0.006
+"Orbital area, ventrolateral part, layer 2/3",0.233,0.25,0.483
+"Temporal association areas, layer 5",0.482,0.439,0.921
+Bed nucleus of the accessory olfactory tract,0.014,0.012,0.026000000000000002
+"Anterior cingulate area, ventral part, layer 2/3",0.198,0.202,0.4
+Magnocellular nucleus,0.162,0.17400000000000002,0.336
+stria terminalis,0.139,0.151,0.29000000000000004
+"Basolateral amygdalar nucleus, anterior part",0.403,0.339,0.742
+"Prelimbic area, layer 2/3",0.167,0.167,0.334
+"Primary visual area, layer 6b",0.08600000000000001,0.093,0.179
+Magnocellular reticular nucleus,0.289,0.271,0.56
+Septofimbrial nucleus,0.212,0.257,0.469
+"Basolateral amygdalar nucleus, posterior part",0.369,0.372,0.741
+Midbrain,3.168,3.358,6.526
+"Agranular insular area, posterior part, layer 6a",0.215,0.20600000000000002,0.42100000000000004
+Supragenual nucleus,0.009000000000000001,0.013000000000000001,0.022000000000000002
+"Primary motor area, Layer 1",0.646,0.623,1.2690000000000001
+Subgeniculate nucleus,0.011,0.013000000000000001,0.024
+Suprageniculate nucleus,0.096,0.095,0.191
+superior cerebelar peduncles,0.20700000000000002,0.20400000000000001,0.41100000000000003
+"Basomedial amygdalar nucleus, anterior part",0.41100000000000003,0.401,0.812
+"Agranular insular area, dorsal part, layer 2/3",0.42,0.466,0.886
+"Retrosplenial area, dorsal part, layer 6b",0.023,0.019,0.041999999999999996
+Accessory supraoptic group,0.001,0.002,0.003
+Septohippocampal nucleus,0.014,0.016,0.03
+"Basomedial amygdalar nucleus, posterior part",0.35100000000000003,0.372,0.7230000000000001
+"Perirhinal area, layer 6a",0.028,0.035,0.063
+superior colliculus commissure,0.015,0.022,0.037
+Subfornical organ,0.008,0.017,0.025
+Substantia innominata,1.522,1.471,2.9930000000000003
+"Agranular insular area, posterior part, layer 5",0.386,0.364,0.75
+Subparaventricular zone,0.07200000000000001,0.046,0.11800000000000001
+supraoptic commissures,0.014,0.016,0.03
+Subceruleus nucleus,0.011,0.015,0.026
+Bed nuclei of the stria terminalis,0.678,0.687,1.3650000000000002
+Medulla,2.5,2.741,5.241
+"Agranular insular area, posterior part, layer 6b",0.01,0.009000000000000001,0.019000000000000003
+Preparasubthalamic nucleus,0.007,0.008,0.015
+Sublaterodorsal nucleus,0.02,0.016,0.036000000000000004
+Mediodorsal nucleus of thalamus,0.6910000000000001,0.672,1.363
+"Prelimbic area, layer 5",0.47000000000000003,0.463,0.933
+Parasubthalamic nucleus,0.07200000000000001,0.077,0.14900000000000002
+Submedial nucleus of the thalamus,0.147,0.163,0.31
+"Perirhinal area, layer 6b",0.008,0.008,0.016
+Infracerebellar nucleus,0.027,0.025,0.052000000000000005
+"Substantia nigra, compact part",0.10300000000000001,0.093,0.196
+"Posterolateral visual area, layer 6a",0.041,0.043000000000000003,0.084
+cuneate fascicle,0.007,0.01,0.017
+"Substantia nigra, reticular part",0.777,0.722,1.499
+Field CA1,5.061,4.947,10.008
+Supraoptic nucleus,0.018000000000000002,0.025,0.043000000000000003
+"Posterolateral visual area, layer 6b",0.004,0.005,0.009000000000000001
+ventral tegmental decussation,0.013000000000000001,0.024,0.037000000000000005
+"Anteromedial visual area, layer 4",0.03,0.024,0.054
+Medial amygdalar nucleus,1.061,0.995,2.056
+"Orbital area, lateral part, layer 2/3",0.279,0.255,0.534
+vestibular nerve,0.109,0.128,0.237
+"Subparafascicular nucleus, magnocellular part",0.033,0.033,0.066
+"Lateral visual area, layer 1",0.092,0.081,0.173
+"Subparafascicular nucleus, parvicellular part",0.089,0.06,0.149
+Field CA2,0.22,0.219,0.439
+Ectorhinal area/Layer 2/3,0.231,0.227,0.458
+medial corticohypothalamic tract,0.005,0.005,0.01
+"Spinal nucleus of the trigeminal, caudal part",0.837,0.8210000000000001,1.658
+"Retrosplenial area, ventral part, layer 2/3",0.539,0.489,1.028
+"Anteromedial visual area, layer 5",0.113,0.101,0.21400000000000002
+"Retrosplenial area, dorsal part, layer 2/3",0.463,0.452,0.915
+columns of the fornix,0.14200000000000002,0.14400000000000002,0.28600000000000003
+"Spinal nucleus of the trigeminal, interpolar part",0.96,0.923,1.883
+"Orbital area, lateral part, layer 6a",0.225,0.231,0.456
+"Anteromedial visual area, layer 6b",0.014,0.015,0.028999999999999998
+"Retrosplenial area, dorsal part, layer 1",0.381,0.403,0.784
+dorsal hippocampal commissure,0.46,0.47000000000000003,0.93
+"Spinal nucleus of the trigeminal, oral part",0.517,0.527,1.044
+"Orbital area, lateral part, layer 1",0.169,0.157,0.326
+ventral hippocampal commissure,0.029,0.041,0.07
+"Primary somatosensory area, upper limb, layer 1",0.245,0.264,0.509
+"Basolateral amygdalar nucleus, ventral part",0.199,0.215,0.41400000000000003
+Median preoptic nucleus,0.002,0.032,0.034
+"Posterior auditory area, layer 6b",0.007,0.008,0.015
+Midbrain trigeminal nucleus,0.006,0.007,0.013000000000000001
+"Primary somatosensory area, trunk, layer 6b",0.021,0.02,0.041
+Field CA3,3.11,3.0140000000000002,6.1240000000000006
+alveus,0.6910000000000001,0.657,1.348
+"posteromedial visual area, layer 6b",0.018000000000000002,0.02,0.038000000000000006
+Subthalamic nucleus,0.10300000000000001,0.112,0.21500000000000002
+Striatum,1.322,1.413,2.7350000000000003
+"Primary somatosensory area, lower limb, layer 6a",0.3,0.228,0.528
+brachium of the inferior colliculus,0.124,0.122,0.246
+Medial habenula,0.145,0.124,0.269
+"Orbital area, medial part, layer 1",0.149,0.269,0.41800000000000004
+"Orbital area, lateral part, layer 6b",0.011,0.023,0.034
+"posteromedial visual area, layer 4",0.044,0.044,0.088
+Subiculum,1.096,0.991,2.087
+dorsal acoustic stria,0.006,0.007,0.013000000000000001
+Main olfactory bulb,3.614,3.855,7.468999999999999
+"Primary somatosensory area, lower limb, layer 6b",0.021,0.019,0.04
+Cerebellum,0.397,0.403,0.8
+Medial preoptic nucleus,0.234,0.216,0.45
+"Ventral auditory area, layer 6a",0.137,0.14100000000000001,0.278
+Medial preoptic area,0.259,0.296,0.5549999999999999
+Supramammillary nucleus,0.11900000000000001,0.139,0.258
+"Entorhinal area, medial part, dorsal zone, layer 1",0.618,0.618,1.236
+"Dorsal auditory area, layer 1",0.09,0.091,0.181
+dorsal fornix,0.004,0.02,0.024
+Medial pretectal area,0.024,0.023,0.047
+Supratrigeminal nucleus,0.16,0.10200000000000001,0.262
+dorsal limb,0.06,0.077,0.137
+"Perirhinal area, layer 1",0.121,0.117,0.238
+"Retrosplenial area, ventral part, layer 1",0.328,0.556,0.8840000000000001
+"Entorhinal area, medial part, dorsal zone, layer 2",0.619,0.63,1.249
+"Central amygdalar nucleus, capsular part",0.14100000000000001,0.158,0.29900000000000004
+Thalamus,0.446,0.424,0.87
+"Central amygdalar nucleus, lateral part",0.134,0.14,0.274
+dorsal spinocerebellar tract,0.075,0.073,0.148
+"Infralimbic area, layer 2/3",0.083,0.075,0.158
+"Primary somatosensory area, nose, layer 1",0.199,0.198,0.397
+"Central amygdalar nucleus, medial part",0.356,0.385,0.741
+Medial septal nucleus,0.121,0.277,0.398
+"posteromedial visual area, layer 5",0.14100000000000001,0.155,0.29600000000000004
+Postpiriform transition area,0.706,0.6970000000000001,1.403
+"Lateral visual area, layer 4",0.092,0.083,0.175
+Tegmental reticular nucleus,0.34400000000000003,0.341,0.685
+Central lateral nucleus of the thalamus,0.163,0.181,0.344
+Accessory facial motor nucleus,0.002,0.002,0.004
+"Primary somatosensory area, upper limb, layer 4",0.265,0.233,0.498
+external capsule,0.455,0.44,0.895
+Nucleus of the brachium of the inferior colliculus,0.043000000000000003,0.042,0.085
+Triangular nucleus of septum,0.156,0.191,0.347
+"Orbital area, medial part, layer 2/3",0.134,0.148,0.28200000000000003
+Claustrum,0.267,0.264,0.531
+Nucleus of Darkschewitsch,0.054,0.05,0.10400000000000001
+"Anterior cingulate area, ventral part, layer 1",0.17200000000000001,0.312,0.484
+"Retrosplenial area, ventral part, layer 6a",0.327,0.316,0.643
+Central linear nucleus raphe,0.03,0.052000000000000005,0.082
+"Primary visual area, layer 1",0.641,0.578,1.2189999999999999
+fasciculus retroflexus,0.083,0.083,0.166
+Diagonal band nucleus,0.343,0.387,0.73
+"Taenia tecta, dorsal part",0.318,0.362,0.6799999999999999
+"Ventral auditory area, layer 6b",0.016,0.022,0.038
+Central medial nucleus of the thalamus,0.1,0.159,0.259
+"Dorsal auditory area, layer 2/3",0.136,0.14400000000000002,0.28
+"Anterolateral visual area, layer 6a",0.06,0.049,0.109
+fimbria,0.745,0.711,1.456
+Nucleus incertus,0.044,0.067,0.111
+"Taenia tecta, ventral part",0.341,0.34400000000000003,0.685
+"Orbital area, ventrolateral part, layer 6a",0.101,0.092,0.193
+Subparafascicular area,0.054,0.08600000000000001,0.14
+"Retrosplenial area, dorsal part, layer 5",0.547,0.577,1.124
+habenular commissure,0.014,0.022,0.036
+Nucleus of the lateral lemniscus,0.384,0.357,0.741
+"Lateral visual area, layer 5",0.171,0.148,0.319
+Tuberal nucleus,0.274,0.27,0.544
+Cuneiform nucleus,0.28,0.281,0.561
+"Orbital area, medial part, layer 5",0.2,0.225,0.42500000000000004
+Motor nucleus of trigeminal,0.163,0.179,0.34199999999999997
+"Retrosplenial area, ventral part, layer 6b",0.021,0.024,0.045
+"Primary somatosensory area, upper limb, layer 5",0.292,0.313,0.605
+Nucleus of the optic tract,0.092,0.10400000000000001,0.196
+Ventral anterior-lateral complex of the thalamus,0.393,0.384,0.777
+"Orbital area, lateral part, layer 5",0.555,0.555,1.11
+"Dentate gyrus, granule cell layer",0.8240000000000001,0.761,1.585
+inferior colliculus commissure,0.007,0.011,0.018
+Nucleus of the posterior commissure,0.148,0.149,0.297
+"Gustatory areas, layer 6a",0.211,0.195,0.406
+"Cortical amygdalar area, anterior part",0.387,0.395,0.782
+Nucleus of the trapezoid body,0.082,0.08,0.162
+"Posterior auditory area, layer 2/3",0.07100000000000001,0.062,0.133
+"Primary motor area, Layer 5",1.51,1.468,2.9779999999999998
+"Anterolateral visual area, layer 6b",0.021,0.016,0.037000000000000005
+Nucleus of the solitary tract,0.40800000000000003,0.437,0.845
+Abducens nucleus,0.02,0.016,0.036000000000000004
+"Primary somatosensory area, nose, layer 4",0.254,0.262,0.516
+"Cortical amygdalar area, posterior part, lateral zone",0.619,0.621,1.24
+"Secondary motor area, layer 1",1.1400000000000001,1.118,2.258
+"Primary somatosensory area, mouth, layer 2/3",0.738,0.738,1.476
+lateral lemniscus,0.428,0.40900000000000003,0.837
+Facial motor nucleus,0.458,0.455,0.913
+"Gustatory areas, layer 6b",0.022,0.021,0.043
+"Cortical amygdalar area, posterior part, medial zone",0.679,0.651,1.33
+"Entorhinal area, medial part, dorsal zone, layer 3",0.506,0.493,0.999
+"lateral olfactory tract, body",0.468,0.456,0.924
+"Frontal pole, layer 2/3",0.131,0.108,0.239
+"Primary somatosensory area, trunk, layer 2/3",0.187,0.196,0.383
+"Retrosplenial area, lateral agranular part, layer 1",0.23,0.24,0.47
+Caudoputamen,12.784,12.508000000000001,25.292
+mammillary peduncle,0.017,0.015,0.032
+"Agranular insular area, ventral part, layer 6a",0.091,0.088,0.179
+"Dorsal auditory area, layer 4",0.065,0.058,0.123
+Superior central nucleus raphe,0.276,0.343,0.619
+"Orbital area, ventrolateral part, layer 6b",0.003,0.004,0.007
+mammillotegmental tract,0.032,0.032,0.064
+Ventral medial nucleus of the thalamus,0.47600000000000003,0.45,0.926
+"Retrosplenial area, ventral part, layer 5",0.807,0.8180000000000001,1.625
+Ventrolateral preoptic nucleus,0.039,0.027,0.066
+mammillothalamic tract,0.058,0.066,0.124
+"Perirhinal area, layer 5",0.084,0.08,0.164
+Ventromedial hypothalamic nucleus,0.28800000000000003,0.28200000000000003,0.5700000000000001
+"Agranular insular area, ventral part, layer 2/3",0.29,0.302,0.592
+"Posterior auditory area, layer 1",0.054,0.048,0.10200000000000001
+medial lemniscus,0.354,0.333,0.687
+Olfactory areas,1.3800000000000001,1.3760000000000001,2.7560000000000002
+"Agranular insular area, ventral part, layer 6b",0.001,0.001,0.002
+"Primary somatosensory area, nose, layer 5",0.28600000000000003,0.278,0.5640000000000001
+Cortical subplate,0.185,0.185,0.37
+"Agranular insular area, ventral part, layer 1",0.111,0.129,0.24
+Olivary pretectal nucleus,0.03,0.034,0.064
+"Infralimbic area, layer 1",0.044,0.101,0.14500000000000002
+Cuneate nucleus,0.152,0.17300000000000001,0.325
+Ventral posterolateral nucleus of the thalamus,0.426,0.388,0.8140000000000001
+"Primary visual area, layer 4",0.502,0.49,0.992
+"Ventral posterolateral nucleus of the thalamus, parvicellular part",0.053,0.044,0.097
+"Entorhinal area, medial part, dorsal zone, layer 5",0.46900000000000003,0.457,0.926
+arbor vitae,3.344,3.317,6.661
+"Temporal association areas, layer 6a",0.262,0.234,0.496
+"Medial mammillary nucleus, median part",0.021,0.045,0.066
+Ventral posteromedial nucleus of the thalamus,0.808,0.864,1.6720000000000002
+"Primary auditory area, layer 1",0.193,0.176,0.369
+"Ventral posteromedial nucleus of the thalamus, parvicellular part",0.132,0.11,0.242
+"Entorhinal area, medial part, dorsal zone, layer 6",0.335,0.314,0.649
+cerebellar commissure,0.03,0.032,0.062
+Ventral tegmental area,0.246,0.211,0.45699999999999996
+"Posterolateral visual area, layer 1",0.096,0.099,0.195
+principal mammillary tract,0.02,0.019,0.039
+Olfactory tubercle,1.956,1.977,3.933
+"Ventral auditory area, layer 2/3",0.189,0.203,0.392
+Ventral tegmental nucleus,0.017,0.02,0.037000000000000005
+"Posterior auditory area, layer 4",0.041,0.039,0.08
+Nucleus x,0.024,0.027,0.051000000000000004
+"Secondary motor area, layer 5",2.253,2.043,4.296
+Pons,1.719,1.94,3.659
+"Anterior cingulate area, ventral part, layer 5",0.492,0.542,1.034
+Hypoglossal nucleus,0.107,0.139,0.246
+"Retrosplenial area, lateral agranular part, layer 5",0.363,0.34900000000000003,0.712
+"Primary visual area, layer 5",0.786,0.768,1.554
+Posterior amygdalar nucleus,0.505,0.492,0.997
+Nucleus y,0.013000000000000001,0.01,0.023
+"Agranular insular area, dorsal part, layer 6a",0.383,0.381,0.764
+corticospinal tract,0.047,0.047,0.094
+"Temporal association areas, layer 6b",0.042,0.03,0.07200000000000001
+Piriform-amygdalar area,0.631,0.638,1.2690000000000001
+"Posterior auditory area, layer 5",0.093,0.089,0.182
+spinal tract of the trigeminal nerve,0.848,0.8210000000000001,1.669
+Periaqueductal gray,1.995,2.058,4.053
+Zona incerta,0.8170000000000001,0.808,1.625
+facial nerve,0.041,0.022,0.063
+"Agranular insular area, ventral part, layer 5",0.35100000000000003,0.359,0.71
+stria medullaris,0.137,0.128,0.265
+Pallidum,0.595,0.578,1.173
+Fields of Forel,0.129,0.14200000000000002,0.271
+"posteromedial visual area, layer 1",0.091,0.079,0.16999999999999998
+"Supplemental somatosensory area, layer 2/3",1.061,0.971,2.032
+"Anterior cingulate area, ventral part, 6a",0.176,0.192,0.368
+"Inferior colliculus, central nucleus",0.549,0.56,1.109
+superior cerebellar peduncle decussation,0.012,0.025,0.037000000000000005
+Dorsal peduncular area,0.23800000000000002,0.24,0.478
+"Primary auditory area, layer 4",0.123,0.11,0.23299999999999998
+"Anterior cingulate area, ventral part, 6b",0.029,0.026000000000000002,0.05500000000000001
+"Inferior colliculus, dorsal nucleus",0.6,0.641,1.241
+"Primary visual area, layer 2/3",1.0130000000000001,0.9560000000000001,1.9690000000000003
+"Infralimbic area, layer 5",0.136,0.14,0.276
+"Inferior colliculus, external nucleus",1.02,0.985,2.005
+Dorsomedial nucleus of the hypothalamus,0.20700000000000002,0.192,0.399
+"Agranular insular area, dorsal part, layer 6b",0.019,0.025,0.044
+oculomotor nerve,0.004,0.006,0.01
+"Superior colliculus, zonal layer",0.133,0.153,0.28600000000000003
+Ectorhinal area/Layer 1,0.124,0.148,0.272
+"Primary somatosensory area, nose, layer 2/3",0.32,0.307,0.627
+Dorsal motor nucleus of the vagus nerve,0.078,0.09,0.16799999999999998
+trapezoid body,0.157,0.177,0.33399999999999996
+"Superior colliculus, superficial gray layer",0.536,0.5710000000000001,1.1070000000000002
+Parasubiculum,0.47700000000000004,0.451,0.928
+"Primary motor area, Layer 6a",1.331,1.337,2.668
+Dentate nucleus,0.169,0.152,0.321
+"Primary auditory area, layer 5",0.34500000000000003,0.313,0.658
+optic nerve,0.04,0.043000000000000003,0.083
+"Visceral area, layer 6b",0.021,0.02,0.041
+uncinate fascicle,0.043000000000000003,0.047,0.09
+"Superior colliculus, optic layer",0.291,0.312,0.603
+Parvicellular reticular nucleus,1.193,1.162,2.355
+"Primary somatosensory area, upper limb, layer 2/3",0.518,0.47600000000000003,0.994
+"Visceral area, layer 6a",0.271,0.265,0.536
+Parasolitary nucleus,0.017,0.013000000000000001,0.030000000000000002
+"Supplemental somatosensory area, layer 6a",0.975,0.93,1.905
+rubrospinal tract,0.28800000000000003,0.28500000000000003,0.5730000000000001
+ventral spinocerebellar tract,0.153,0.149,0.302
+Parabrachial nucleus,0.47800000000000004,0.464,0.9420000000000001
+"Posterolateral visual area, layer 4",0.016,0.011,0.027
+Dorsal nucleus raphe,0.033,0.124,0.157
+"Supplemental somatosensory area, layer 1",0.637,0.598,1.2349999999999999
+Parabigeminal nucleus,0.016,0.021,0.037000000000000005
+"Primary somatosensory area, mouth, layer 1",0.384,0.377,0.761
+Dorsal tegmental nucleus,0.056,0.061,0.11699999999999999
+"Primary motor area, Layer 6b",0.098,0.088,0.186
+amygdalar capsule,0.084,0.088,0.172
+"Perirhinal area, layer 2/3",0.168,0.169,0.337
+"Primary somatosensory area, nose, layer 6a",0.451,0.379,0.8300000000000001
+"Supplemental somatosensory area, layer 6b",0.10200000000000001,0.08600000000000001,0.188
+"Visceral area, layer 1",0.169,0.156,0.325
+Pontine central gray,0.26,0.229,0.489
+"anterior commissure, olfactory limb",0.382,0.376,0.758
+"Posterolateral visual area, layer 5",0.101,0.115,0.21600000000000003
+External cuneate nucleus,0.105,0.105,0.21
+"Anterolateral visual area, layer 2/3",0.091,0.098,0.189
+"Retrosplenial area, lateral agranular part, layer 6a",0.203,0.20400000000000001,0.40700000000000003
+Paracentral nucleus,0.108,0.117,0.225
+"anterior commissure, temporal limb",0.115,0.111,0.226
+"Orbital area, medial part, layer 6a",0.088,0.08,0.16799999999999998
+trochlear nerve,0.004,0.004,0.008
+Lingula (I),0.049,0.057,0.10600000000000001
+Posterodorsal preoptic nucleus,0.004,0.005,0.009000000000000001
+brachium of the superior colliculus,0.085,0.08600000000000001,0.171
+"Anterior cingulate area, dorsal part, layer 6a",0.326,0.363,0.6890000000000001
+cerebal peduncle,0.465,0.455,0.92
+"Anterior cingulate area, dorsal part, layer 6b",0.01,0.02,0.03
+"Primary somatosensory area, nose, layer 6b",0.025,0.025,0.05
+Parafascicular nucleus,0.23800000000000002,0.264,0.502
+Pontine gray,0.485,0.48,0.965
+"Anterior cingulate area, dorsal part, layer 1",0.20700000000000002,0.353,0.56
+Declive (VI),1.472,1.691,3.1630000000000003
+"Nucleus ambiguus, dorsal division",0.013000000000000001,0.015,0.028
+cingulum bundle,0.655,0.553,1.2080000000000002
+"Primary motor area, Layer 2/3",1.8860000000000001,1.653,3.539
+Folium-tuber vermis (VII),0.494,0.534,1.028
+"Primary somatosensory area, upper limb, layer 6a",0.505,0.429,0.9339999999999999
+Posterior hypothalamic nucleus,0.328,0.363,0.6910000000000001
+vomeronasal nerve,0.008,0.006,0.014
+"Primary somatosensory area, mouth, layer 4",0.461,0.463,0.924
+Pyramus (VIII),0.515,0.622,1.137
+"Endopiriform nucleus, dorsal part",0.89,0.894,1.784
+"Primary auditory area, layer 6a",0.134,0.134,0.268
+"Lateral reticular nucleus, magnocellular part",0.27,0.264,0.534
+"corpus callosum, anterior forceps",0.418,0.417,0.835
+Uvula (IX),0.918,1.1500000000000001,2.068
+"Ventral auditory area, layer 1",0.131,0.135,0.266
+Piriform area,5.9,5.776,11.676
+"Secondary motor area, layer 2/3",1.8860000000000001,1.848,3.734
+"Lateral reticular nucleus, parvicellular part",0.03,0.031,0.061
+"corpus callosum, extreme capsule",0.057,0.048,0.10500000000000001
+"Retrosplenial area, lateral agranular part, layer 2/3",0.31,0.309,0.619
+"Endopiriform nucleus, ventral part",0.485,0.47900000000000004,0.964
+Nodulus (X),0.666,0.768,1.4340000000000002
+"Orbital area, ventrolateral part, layer 1",0.185,0.199,0.384
+"Paragigantocellular reticular nucleus, dorsal part",0.11800000000000001,0.101,0.21900000000000003
+"corpus callosum, posterior forceps",0.675,0.666,1.3410000000000002
+"Lateral visual area, layer 2/3",0.16,0.135,0.29500000000000004
+"Primary somatosensory area, mouth, layer 5",0.546,0.587,1.133
+Lobule II,0.613,0.626,1.2389999999999999
+Ectorhinal area/Layer 6a,0.195,0.183,0.378
+"Paragigantocellular reticular nucleus, lateral part",0.381,0.35100000000000003,0.732
+Dorsal premammillary nucleus,0.07200000000000001,0.069,0.14100000000000001
+"Primary somatosensory area, barrel field, layer 1",0.403,0.372,0.775
+Fasciola cinerea,0.031,0.031,0.062
+Lobule III,1.179,1.414,2.593
+"corpus callosum, splenium",0.329,0.329,0.658
+Ectorhinal area/Layer 5,0.267,0.254,0.521
+Fastigial nucleus,0.245,0.26,0.505
+"Ventral auditory area, layer 4",0.109,0.10400000000000001,0.21300000000000002
+"Agranular insular area, dorsal part, layer 1",0.224,0.23,0.454
+root,1.677,1.864,3.5410000000000004
+Fundus of striatum,0.23800000000000002,0.20400000000000001,0.44200000000000006
+Ventral premammillary nucleus,0.111,0.096,0.20700000000000002
+"Primary auditory area, layer 6b",0.017,0.019,0.036000000000000004
+"Primary somatosensory area, trunk, layer 1",0.098,0.093,0.191
+Simple lobule,2.845,2.587,5.432
+fiber tracts,0.777,0.772,1.549
+"Visceral area, layer 4",0.1,0.096,0.196
+"Anterior cingulate area, dorsal part, layer 5",0.54,0.529,1.069
+olfactory nerve layer of main olfactory bulb,0.503,0.5660000000000001,1.069
+Posterior complex of the thalamus,0.642,0.599,1.241
+"Secondary motor area, layer 6a",0.978,1.173,2.151
+"Globus pallidus, external segment",0.803,0.759,1.562
+"Ventral auditory area, layer 5",0.303,0.295,0.598
+Paramedian lobule,2.477,2.345,4.822
+"Primary somatosensory area, upper limb, layer 6b",0.041,0.045,0.086
+Posterior limiting nucleus of the thalamus,0.1,0.092,0.192
+"Primary somatosensory area, lower limb, layer 1",0.165,0.127,0.29200000000000004
+"Globus pallidus, internal segment",0.20500000000000002,0.198,0.403
+Copula pyramidis,1.258,1.057,2.315
+"Supplemental somatosensory area, layer 4",0.59,0.544,1.134
+Postsubiculum,0.535,0.509,1.044
+"Primary somatosensory area, barrel field, layer 6a",0.637,0.605,1.242
+Gracile nucleus,0.038,0.042,0.08
+Paraflocculus,2.914,2.7520000000000002,5.666
+crossed tectospinal pathway,0.25,0.243,0.493
+Peripeduncular nucleus,0.036000000000000004,0.035,0.07100000000000001
+Ectorhinal area/Layer 6b,0.033,0.025,0.058
+"Anteromedial visual area, layer 6a",0.061,0.058,0.119
+"Primary somatosensory area, barrel field, layer 4",0.641,0.599,1.24
+Gigantocellular reticular nucleus,1.262,1.303,2.565
+Flocculus,0.655,0.658,1.3130000000000002
+Pedunculopontine nucleus,0.464,0.39,0.8540000000000001
+"Infralimbic area, layer 6a",0.10200000000000001,0.095,0.197
+Crus 1,2.82,2.725,5.545
+"Visceral area, layer 5",0.355,0.353,0.708
+doral tegmental decussation,0.001,0.003,0.004
+Posterior pretectal nucleus,0.07100000000000001,0.07100000000000001,0.14200000000000002
+"Primary somatosensory area, barrel field, layer 6b",0.059000000000000004,0.07100000000000001,0.13
+Crus 2,2.481,2.52,5.0009999999999994
+"Anteromedial visual area, layer 2/3",0.114,0.089,0.203
+Parapyramidal nucleus,0.056,0.049,0.10500000000000001
+"Primary somatosensory area, barrel field, layer 5",0.635,0.5660000000000001,1.201
+"Medial geniculate complex, dorsal part",0.081,0.076,0.157
+"Anterolateral visual area, layer 1",0.056,0.063,0.119
+Perireunensis nucleus,0.075,0.082,0.157
+"Medial geniculate complex, ventral part",0.125,0.129,0.254
+"Infralimbic area, layer 6b",0.003,0.004,0.007
+Presubiculum,0.47100000000000003,0.445,0.916
+"Secondary motor area, layer 6b",0.037,0.039,0.076
+"Primary somatosensory area, trunk, layer 4",0.063,0.068,0.131
+"Medial geniculate complex, medial part",0.115,0.107,0.222
+Hippocampal formation,0.195,0.209,0.404
+"Supplemental somatosensory area, layer 5",1.065,0.997,2.062
+Lobules IV-V,3.079,3.399,6.478
+external medullary lamina of the thalamus,0.053,0.052000000000000005,0.10500000000000001
+"Pontine reticular nucleus, caudal part",1.21,1.218,2.428
+"Primary somatosensory area, lower limb, layer 4",0.131,0.153,0.28400000000000003
+"Anteromedial nucleus, dorsal part",0.135,0.105,0.24
+Hypothalamus,1.359,1.421,2.7800000000000002
+"Medullary reticular nucleus, dorsal part",0.533,0.516,1.049
+"Agranular insular area, dorsal part, layer 5",0.753,0.748,1.501
+"Primary somatosensory area, mouth, layer 6a",0.872,0.792,1.6640000000000001
+"Anteromedial nucleus, ventral part",0.095,0.07,0.165
+Intercalated amygdalar nucleus,0.10300000000000001,0.079,0.182
+"Visceral area, layer 2/3",0.275,0.269,0.544
+"Medullary reticular nucleus, ventral part",0.421,0.456,0.877
+genu of corpus callosum,0.403,0.392,0.795
+Parastrial nucleus,0.053,0.05,0.10300000000000001
+"Primary somatosensory area, trunk, layer 5",0.191,0.168,0.359
+Interanterodorsal nucleus of the thalamus,0.061,0.067,0.128
+"Anterolateral visual area, layer 4",0.053,0.049,0.10200000000000001
+genu of the facial nerve,0.019,0.014,0.033
+Interanteromedial nucleus of the thalamus,0.022,0.02,0.041999999999999996
+"Entorhinal area, lateral part, layer 1",0.494,0.46900000000000003,0.9630000000000001
+inferior cerebellar peduncle,0.372,0.363,0.735
+"Orbital area, ventrolateral part, layer 5",0.276,0.28300000000000003,0.559
+"Tuberomammillary nucleus, dorsal part",0.026000000000000002,0.024,0.05
+"Temporal association areas, layer 2/3",0.377,0.329,0.706
+"Primary somatosensory area, lower limb, layer 5",0.211,0.304,0.515
+"Nucleus of the lateral olfactory tract, layer 3",0.037,0.029,0.066
+Median eminence,0.029,0.053,0.082
+"Dentate gyrus, molecular layer",2.233,2.071,4.304
+"Dentate gyrus, polymorph layer",0.324,0.295,0.619
+"Primary somatosensory area, unassigned, layer 1",0.084,0.067,0.15100000000000002
+"Primary somatosensory area, unassigned, layer 2/3",0.17500000000000002,0.138,0.31300000000000006
+"Primary somatosensory area, unassigned, layer 4",0.075,0.08,0.155
+"Primary somatosensory area, unassigned, layer 5",0.11900000000000001,0.12,0.239
+"Primary somatosensory area, unassigned, layer 6a",0.164,0.158,0.322
+"Primary somatosensory area, unassigned, layer 6b",0.016,0.015,0.031
+"Anterior area, layer 1",0.10200000000000001,0.117,0.21900000000000003
+"Anterior area, layer 2/3",0.209,0.20700000000000002,0.41600000000000004
+"Anterior area, layer 4",0.089,0.073,0.16199999999999998
+"Anterior area, layer 5",0.2,0.16,0.36
+"Anterior area, layer 6a",0.11,0.094,0.20400000000000001
+"Anterior area, layer 6b",0.022,0.023,0.045
+"Laterointermediate area, layer 1",0.036000000000000004,0.032,0.068
+"Laterointermediate area, layer 2/3",0.06,0.054,0.11399999999999999
+"Laterointermediate area, layer 4",0.03,0.031,0.061
+"Laterointermediate area, layer 5",0.078,0.069,0.14700000000000002
+"Laterointermediate area, layer 6a",0.04,0.032,0.07200000000000001
+"Laterointermediate area, layer 6b",0.008,0.009000000000000001,0.017
+"Rostrolateral area, layer 1",0.088,0.081,0.16899999999999998
+"Rostrolateral area, layer 2/3",0.148,0.134,0.28200000000000003
+"Rostrolateral area, layer 4",0.075,0.07,0.14500000000000002
+"Rostrolateral area, layer 5",0.111,0.116,0.227
+"Rostrolateral area, layer 6a",0.073,0.079,0.152
+"Rostrolateral area, layer 6b",0.011,0.016,0.027
+"Postrhinal area, layer 1",0.113,0.117,0.23
+"Postrhinal area, layer 2/3",0.178,0.163,0.34099999999999997
+"Postrhinal area, layer 4",0.033,0.03,0.063
+"Postrhinal area, layer 5",0.17300000000000001,0.185,0.358
+"Postrhinal area, layer 6a",0.082,0.09,0.172
+"Postrhinal area, layer 6b",0.017,0.019,0.036000000000000004
+Prosubiculum,0.58,0.562,1.142
+Area prostriata,0.181,0.183,0.364
+supra-callosal cerebral white matter,0.56,0.501,1.061
+"corpus callosum, body",1.458,1.252,2.71
+optic radiation,0.865,0.867,1.732
+auditory radiation,0.22,0.198,0.41800000000000004
+commissural branch of stria terminalis,0.017,0.018000000000000002,0.035
+"Dorsal part of the lateral geniculate complex, shell",0.10400000000000001,0.097,0.201
+"Dorsal part of the lateral geniculate complex, core",0.211,0.203,0.41400000000000003
+"Dorsal part of the lateral geniculate complex, ipsilateral zone",0.043000000000000003,0.043000000000000003,0.08600000000000001
+"Frontal pole, layer 5",0.176,0.17500000000000002,0.351
+"Frontal pole, layer 6a",0.04,0.048,0.088
+"Frontal pole, layer 6b",0.001,0.001,0.002
+"Orbital area, medial part, layer 6b",0.004,0.002,0.006
+Retroparafascicular nucleus,0.027,0.033,0.06
+Medial accesory oculomotor nucleus,0.01,0.006,0.016
+Peritrigeminal zone,0.158,0.162,0.32
+Accessory trigeminal nucleus,0.005,0.007,0.012
+Parvicellular motor 5 nucleus,0.029,0.033,0.062
+Intertrigeminal nucleus,0.02,0.028,0.048
+Ethmoid nucleus of the thalamus,0.129,0.112,0.241
+Xiphoid thalamic nucleus,0.007,0.07100000000000001,0.07800000000000001
+Posterior intralaminar thalamic nucleus,0.101,0.091,0.192
+Posterior triangular thalamic nucleus,0.131,0.128,0.259
+Intermediate geniculate nucleus,0.01,0.01,0.02
+Ventromedial preoptic nucleus,0.02,0.021,0.041
+Perifornical nucleus,0.113,0.108,0.221
+Hippocampo-amygdalar transition area,0.203,0.2,0.403
+Paratrigeminal nucleus,0.054,0.05,0.10400000000000001
+Vestibulocerebellar nucleus,0.039,0.046,0.08499999999999999
+Subcommissural organ,0.002,0.009000000000000001,0.011000000000000001
+Posterodorsal tegmental nucleus,0.021,0.022,0.043
+"Medial mammillary nucleus, lateral part",0.111,0.11,0.221
+"Medial mammillary nucleus, medial part",0.065,0.07,0.135
+"Medial mammillary nucleus, posterior part",0.011,0.02,0.031
+"Medial mammillary nucleus, dorsal part",0.032,0.044,0.076
+Paratrochlear nucleus,0.011,0.007,0.018
+Paranigral nucleus,0.012,0.011,0.023
+"Interpeduncular nucleus, rostral",0.029,0.051000000000000004,0.08
+"Interpeduncular nucleus, caudal",0.021,0.045,0.066
+"Interpeduncular nucleus, apical",0.004,0.012,0.016
+"Interpeduncular nucleus, lateral",0.029,0.031,0.06
+"Interpeduncular nucleus, intermediate",0.02,0.021,0.041
+"Interpeduncular nucleus, dorsomedial",0.011,0.009000000000000001,0.02
+"Interpeduncular nucleus, dorsolateral",0.015,0.018000000000000002,0.033
+"Interpeduncular nucleus, rostrolateral",0.009000000000000001,0.009000000000000001,0.018000000000000002
+Supraoculomotor periaqueductal gray,0.015,0.018000000000000002,0.033
diff --git a/tests/data/signal/signal_ch000000.tif b/tests/data/signal/signal_ch000000.tif
new file mode 100644
index 00000000..0c3456ce
Binary files /dev/null and b/tests/data/signal/signal_ch000000.tif differ
diff --git a/tests/data/signal/signal_ch000001.tif b/tests/data/signal/signal_ch000001.tif
new file mode 100644
index 00000000..085b2afb
Binary files /dev/null and b/tests/data/signal/signal_ch000001.tif differ
diff --git a/tests/data/signal/signal_ch000002.tif b/tests/data/signal/signal_ch000002.tif
new file mode 100644
index 00000000..a5b65252
Binary files /dev/null and b/tests/data/signal/signal_ch000002.tif differ
diff --git a/tests/data/signal/signal_ch000003.tif b/tests/data/signal/signal_ch000003.tif
new file mode 100644
index 00000000..367b8532
Binary files /dev/null and b/tests/data/signal/signal_ch000003.tif differ
diff --git a/tests/data/signal/signal_ch000004.tif b/tests/data/signal/signal_ch000004.tif
new file mode 100644
index 00000000..81a0d018
Binary files /dev/null and b/tests/data/signal/signal_ch000004.tif differ
diff --git a/tests/data/signal/signal_ch000005.tif b/tests/data/signal/signal_ch000005.tif
new file mode 100644
index 00000000..cb479e8e
Binary files /dev/null and b/tests/data/signal/signal_ch000005.tif differ
diff --git a/tests/data/signal/signal_ch000006.tif b/tests/data/signal/signal_ch000006.tif
new file mode 100644
index 00000000..56161f0d
Binary files /dev/null and b/tests/data/signal/signal_ch000006.tif differ
diff --git a/tests/data/signal/signal_ch000007.tif b/tests/data/signal/signal_ch000007.tif
new file mode 100644
index 00000000..7af83e4d
Binary files /dev/null and b/tests/data/signal/signal_ch000007.tif differ
diff --git a/tests/data/signal/signal_ch000008.tif b/tests/data/signal/signal_ch000008.tif
new file mode 100644
index 00000000..03795ddf
Binary files /dev/null and b/tests/data/signal/signal_ch000008.tif differ
diff --git a/tests/data/signal/signal_ch000009.tif b/tests/data/signal/signal_ch000009.tif
new file mode 100644
index 00000000..9d28134d
Binary files /dev/null and b/tests/data/signal/signal_ch000009.tif differ
diff --git a/tests/data/signal/signal_ch000010.tif b/tests/data/signal/signal_ch000010.tif
new file mode 100644
index 00000000..30aa0b20
Binary files /dev/null and b/tests/data/signal/signal_ch000010.tif differ
diff --git a/tests/data/signal/signal_ch000011.tif b/tests/data/signal/signal_ch000011.tif
new file mode 100644
index 00000000..e361256e
Binary files /dev/null and b/tests/data/signal/signal_ch000011.tif differ
diff --git a/tests/data/signal/signal_ch000012.tif b/tests/data/signal/signal_ch000012.tif
new file mode 100644
index 00000000..da655f39
Binary files /dev/null and b/tests/data/signal/signal_ch000012.tif differ
diff --git a/tests/data/signal/signal_ch000013.tif b/tests/data/signal/signal_ch000013.tif
new file mode 100644
index 00000000..8f176dd4
Binary files /dev/null and b/tests/data/signal/signal_ch000013.tif differ
diff --git a/tests/data/signal/signal_ch000014.tif b/tests/data/signal/signal_ch000014.tif
new file mode 100644
index 00000000..0246e1a8
Binary files /dev/null and b/tests/data/signal/signal_ch000014.tif differ
diff --git a/tests/data/signal/signal_ch000015.tif b/tests/data/signal/signal_ch000015.tif
new file mode 100644
index 00000000..86e44f1f
Binary files /dev/null and b/tests/data/signal/signal_ch000015.tif differ
diff --git a/tests/data/signal/signal_ch000016.tif b/tests/data/signal/signal_ch000016.tif
new file mode 100644
index 00000000..75a9f430
Binary files /dev/null and b/tests/data/signal/signal_ch000016.tif differ
diff --git a/tests/data/signal/signal_ch000017.tif b/tests/data/signal/signal_ch000017.tif
new file mode 100644
index 00000000..dde72ae8
Binary files /dev/null and b/tests/data/signal/signal_ch000017.tif differ
diff --git a/tests/data/signal/signal_ch000018.tif b/tests/data/signal/signal_ch000018.tif
new file mode 100644
index 00000000..e07e823f
Binary files /dev/null and b/tests/data/signal/signal_ch000018.tif differ
diff --git a/tests/data/signal/signal_ch000019.tif b/tests/data/signal/signal_ch000019.tif
new file mode 100644
index 00000000..71353e6e
Binary files /dev/null and b/tests/data/signal/signal_ch000019.tif differ
diff --git a/tests/data/signal/signal_ch000020.tif b/tests/data/signal/signal_ch000020.tif
new file mode 100644
index 00000000..897355b4
Binary files /dev/null and b/tests/data/signal/signal_ch000020.tif differ
diff --git a/tests/data/signal/signal_ch000021.tif b/tests/data/signal/signal_ch000021.tif
new file mode 100644
index 00000000..619851d6
Binary files /dev/null and b/tests/data/signal/signal_ch000021.tif differ
diff --git a/tests/data/signal/signal_ch000022.tif b/tests/data/signal/signal_ch000022.tif
new file mode 100644
index 00000000..95ed5efc
Binary files /dev/null and b/tests/data/signal/signal_ch000022.tif differ
diff --git a/tests/data/signal/signal_ch000023.tif b/tests/data/signal/signal_ch000023.tif
new file mode 100644
index 00000000..96078183
Binary files /dev/null and b/tests/data/signal/signal_ch000023.tif differ
diff --git a/tests/data/signal/signal_ch000024.tif b/tests/data/signal/signal_ch000024.tif
new file mode 100644
index 00000000..a5e63056
Binary files /dev/null and b/tests/data/signal/signal_ch000024.tif differ
diff --git a/tests/data/signal/signal_ch000025.tif b/tests/data/signal/signal_ch000025.tif
new file mode 100644
index 00000000..474232e4
Binary files /dev/null and b/tests/data/signal/signal_ch000025.tif differ
diff --git a/tests/tests/conftest.py b/tests/tests/conftest.py
new file mode 100644
index 00000000..6f29d259
--- /dev/null
+++ b/tests/tests/conftest.py
@@ -0,0 +1,56 @@
+import pathlib
+import sys
+
+import pytest
+from brainglobe_utils.general.config import get_config_obj
+from cellfinder_core.download.cli import main as cellfinder_download
+
+test_data_dir = pathlib.Path(__file__) / ".." / ".." / "data"
+data_dir = test_data_dir / "brain"
+test_output_dir = test_data_dir / "registration_output"
+
+TEST_ATLAS = "allen_2017_100um"
+
+
+def download_atlas(directory):
+ download_args = [
+ "cellfinder_download",
+ "--atlas",
+ TEST_ATLAS,
+ "--install-path",
+ directory,
+ "--no-amend-config",
+ "--no-models",
+ ]
+ sys.argv = download_args
+ cellfinder_download()
+ return directory
+
+
+def generate_test_config(atlas_dir):
+ config = test_data_dir / "config" / "test.conf"
+ config_obj = get_config_obj(config)
+ atlas_conf = config_obj["atlas"]
+ orig_base_directory = atlas_conf["base_folder"]
+
+ with open(config, "r") as in_conf:
+ data = in_conf.readlines()
+ for i, line in enumerate(data):
+ data[i] = line.replace(
+ f"base_folder = '{orig_base_directory}",
+ f"base_folder = '{atlas_dir / 'atlas' / TEST_ATLAS}",
+ )
+ test_config = atlas_dir / "config.conf"
+ with open(test_config, "w") as out_conf:
+ out_conf.writelines(data)
+
+ return test_config
+
+
+@pytest.fixture()
+def test_config_path(tmpdir):
+ print("fixture")
+ atlas_directory = str(tmpdir)
+ download_atlas(atlas_directory)
+ test_config = generate_test_config(atlas_directory)
+ return test_config
diff --git a/tests/tests/test_integration/test_detection.py b/tests/tests/test_integration/test_detection.py
new file mode 100644
index 00000000..e4cd5e26
--- /dev/null
+++ b/tests/tests/test_integration/test_detection.py
@@ -0,0 +1,76 @@
+import os
+import sys
+from math import isclose
+
+import brainglobe_utils.IO.cells as cell_io
+import pytest
+
+from cellfinder.main import main as cellfinder_run
+
+data_dir = os.path.join(
+ os.getcwd(), "tests", "data", "integration", "detection"
+)
+signal_data = os.path.join(data_dir, "crop_planes", "ch0")
+background_data = os.path.join(data_dir, "crop_planes", "ch1")
+cells_validation_xml = os.path.join(data_dir, "cell_classification.xml")
+
+x_pix = "2"
+y_pix = "2"
+z_pix = "5"
+
+DETECTION_TOLERANCE = 2
+
+
+# FIXME: This isn't a very good example
+
+
+@pytest.mark.slow
+def test_detection_full(tmpdir):
+ cellfinder_args = [
+ "cellfinder",
+ "-s",
+ signal_data,
+ "-b",
+ background_data,
+ "-o",
+ str(tmpdir),
+ "-v",
+ z_pix,
+ y_pix,
+ x_pix,
+ "--orientation",
+ "psl",
+ "--n-free-cpus",
+ "0",
+ "--no-register",
+ "--save-planes",
+ ]
+ sys.argv = cellfinder_args
+ cellfinder_run()
+
+ cells_test_xml = tmpdir / "points" / "cell_classification.xml"
+
+ cells_validation = cell_io.get_cells(cells_validation_xml)
+ cells_test = cell_io.get_cells(str(cells_test_xml))
+
+ num_non_cells_validation = sum(
+ [cell.type == 1 for cell in cells_validation]
+ )
+ num_cells_validation = sum([cell.type == 2 for cell in cells_validation])
+
+ num_non_cells_test = sum([cell.type == 1 for cell in cells_test])
+ num_cells_test = sum([cell.type == 2 for cell in cells_test])
+
+ assert isclose(
+ num_non_cells_validation,
+ num_non_cells_test,
+ abs_tol=DETECTION_TOLERANCE,
+ )
+ assert isclose(
+ num_cells_validation, num_cells_test, abs_tol=DETECTION_TOLERANCE
+ )
+ # Check that planes are saved
+ for i in range(2, 30):
+ assert (
+ tmpdir / "processed_planes" / f"plane_{str(i).zfill(4)}.tif"
+ ).exists()
diff --git a/tests/tests/test_integration/test_extract.py b/tests/tests/test_integration/test_extract.py
new file mode 100644
index 00000000..1116ee58
--- /dev/null
+++ b/tests/tests/test_integration/test_extract.py
@@ -0,0 +1,170 @@
+import os
+
+import imio
+import numpy as np
+import pytest
+from brainglobe_utils.cells.cells import Cell
+from brainglobe_utils.general.system import (
+ delete_directory_contents,
+ get_sorted_file_paths,
+)
+from brainglobe_utils.IO.cells import get_cells
+from tifffile import tifffile
+
+import cellfinder.extract.extract_cubes as extract_cubes
+
+data_dir = os.path.join("tests", "data")
+
+signal_data_dir = os.path.join(data_dir, "signal")
+background_data_dir = os.path.join(data_dir, "background")
+xml_path = os.path.join(data_dir, "cube_extract", "cells.xml")
+validate_cubes_dir = os.path.join(data_dir, "cube_extract", "cubes")
+validate_cubes_scale_dir = os.path.join(
+ data_dir, "cube_extract", "cubes_scale"
+)
+
+
+class CubeExtractArgs:
+ def __init__(self, tmpdir):
+ self.cells_file_path = xml_path
+
+ self.cube_width = 50
+ self.cube_height = 50
+ self.cube_depth = 20
+ self.save_empty_cubes = False
+ self.voxel_sizes = [5, 1, 1]
+
+ # get these from parser defaults
+ self.network_voxel_sizes = [5, 1, 1]
+
+ self.n_free_cpus = 0
+ self.n_max_threads = 10
+ self.max_ram = None
+
+ self.paths = Paths(cells_file_path=xml_path, cubes_output_dir=tmpdir)
+
+ @staticmethod
+ def get_plane_paths():
+ return signal_data_dir + background_data_dir
+
+
+class Paths:
+ def __init__(self, cells_file_path=None, cubes_output_dir=None):
+ self.cells_file_path = cells_file_path
+ self.tmp__cubes_output_dir = cubes_output_dir
+
+
+def load_cubes_in_dir(directory):
+ cube_list = os.listdir(directory)
+ cubes = []
+ for file in cube_list:
+ file_path = os.path.join(directory, file)
+ cubes.append(imio.load_any(file_path))
+ return cubes
+
+
+def test_cube_extraction(tmpdir, depth=20):
+ tmpdir = str(tmpdir)
+ args = CubeExtractArgs(tmpdir)
+
+ planes_paths = {}
+ planes_paths[0] = get_sorted_file_paths(
+ signal_data_dir, file_extension="tif"
+ )
+ planes_paths[1] = get_sorted_file_paths(
+ background_data_dir, file_extension="tif"
+ )
+
+ extract_cubes.main(
+ get_cells(args.paths.cells_file_path),
+ args.paths.tmp__cubes_output_dir,
+ planes_paths,
+ args.cube_depth,
+ args.cube_width,
+ args.cube_height,
+ args.voxel_sizes,
+ args.network_voxel_sizes,
+ args.max_ram,
+ args.n_free_cpus,
+ args.save_empty_cubes,
+ )
+
+ validation_cubes = load_cubes_in_dir(validate_cubes_dir)
+ test_cubes = load_cubes_in_dir(tmpdir)
+
+ for idx, test_cube in enumerate(test_cubes):
+ assert (validation_cubes[idx] == test_cube).all()
+
+ delete_directory_contents(tmpdir)
+
+ # test cube scaling
+ args.voxel_sizes = [7.25, 2, 2]
+ args.x_pixel_um = 2
+ args.y_pixel_um = 2
+ args.z_pixel_um = 7.25
+
+ extract_cubes.main(
+ get_cells(args.paths.cells_file_path),
+ args.paths.tmp__cubes_output_dir,
+ planes_paths,
+ args.cube_depth,
+ args.cube_width,
+ args.cube_height,
+ args.voxel_sizes,
+ args.network_voxel_sizes,
+ args.max_ram,
+ args.n_free_cpus,
+ args.save_empty_cubes,
+ )
+
+ validation_cubes_scale = load_cubes_in_dir(validate_cubes_scale_dir)
+ test_cubes = load_cubes_in_dir(tmpdir)
+ for idx, test_cube in enumerate(test_cubes):
+ assert (validation_cubes_scale[idx] == test_cube).all()
+
+ # test edge of data errors
+ cell = Cell("x0y0z10", 2)
+ plane_paths = os.listdir(signal_data_dir)
+ first_plane = tifffile.imread(
+ os.path.join(signal_data_dir, plane_paths[0])
+ )
+ stack_shape = first_plane.shape + (depth,)
+ stacks = {}
+ stacks[0] = np.zeros(stack_shape, dtype=np.uint16)
+ stacks[0][:, :, 0] = first_plane
+
+ for plane in range(1, depth):
+ im_path = os.path.join(signal_data_dir, plane_paths[plane])
+ stacks[0][:, :, plane] = tifffile.imread(im_path)
+
+ cube = extract_cubes.Cube(cell, 0, stacks)
+ assert (cube.data == 0).all()
+
+ cell = Cell("x2500y2500z10", 2)
+ cube = extract_cubes.Cube(cell, 0, stacks)
+ assert (cube.data == 0).all()
+
+ # test insufficient z-planes for a specific cube
+ stacks[0] = stacks[0][:, :, 1:]
+ cube = extract_cubes.Cube(cell, 0, stacks)
+ assert (cube.data == 0).all()
+
+ # test insufficient z-planes for any cube to be extracted at all.
+ delete_directory_contents(tmpdir)
+ # args.z_pixel_um = 0.1
+ args.voxel_sizes[0] = 0.1
+
+ with pytest.raises(extract_cubes.StackSizeError):
+ extract_cubes.main(
+ get_cells(args.paths.cells_file_path),
+ args.paths.tmp__cubes_output_dir,
+ planes_paths,
+ args.cube_depth,
+ args.cube_width,
+ args.cube_height,
+ args.voxel_sizes,
+ args.network_voxel_sizes,
+ args.max_ram,
+ args.n_free_cpus,
+ args.save_empty_cubes,
+ )
diff --git a/tests/tests/test_integration/test_registration.py b/tests/tests/test_integration/test_registration.py
new file mode 100644
index 00000000..cc6d66f7
--- /dev/null
+++ b/tests/tests/test_integration/test_registration.py
@@ -0,0 +1,111 @@
+import os
+import platform
+import sys
+
+import numpy as np
+import pandas as pd
+import pytest
+from imio.load import load_any
+
+from cellfinder.main import main as cellfinder_run
+
+data_dir = os.path.join(
+ os.getcwd(),
+ "tests",
+ "data",
+ "brain",
+)
+
+test_niftyreg_output = os.path.join(
+ os.getcwd(), "tests", "data", "registration_output", platform.system()
+)
+
+x_pix = "40"
+y_pix = "40"
+z_pix = "50"
+
+relative_tolerance = 0.01
+absolute_tolerance = 10
+check_less_precise_pd = 1
+
+
+@pytest.mark.xfail(reason="Issues across machines")
+@pytest.mark.slow
+def test_registration_niftyreg(tmpdir):
+ output_directory = os.path.join(str(tmpdir), "output")
+ cellfinder_args = [
+ "cellfinder",
+ "-s",
+ data_dir,
+ "-b",
+ data_dir,
+ "-o",
+ output_directory,
+ "-v",
+ z_pix,
+ y_pix,
+ x_pix,
+ "--orientation",
+ "psl",
+ "--n-free-cpus",
+ "0",
+ "--atlas",
+ "allen_mouse_100um",
+ "--no-detection",
+ "--no-classification",
+ "--no-analyse",
+ "--no-figures",
+ ]
+
+ sys.argv = cellfinder_args
+ cellfinder_run()
+
+ # none of this testing is ideal, as results seem to vary between systems
+
+ if platform.system() == "Linux":
+ image_list = [
+ "boundaries.tiff",
+ "deformation_field_0.tiff",
+ "deformation_field_1.tiff",
+ "deformation_field_2.tiff",
+ "downsampled.tiff",
+ "downsampled_channel_0.tiff",
+ "downsampled_standard.tiff",
+ "downsampled_standard_channel_0.tiff",
+ "registered_atlas.tiff",
+ "registered_hemispheres.tiff",
+ ]
+ else:
+ image_list = [
+ "boundaries.tiff",
+ "deformation_field_0.tiff",
+ "deformation_field_1.tiff",
+ "deformation_field_2.tiff",
+ "downsampled.tiff",
+ "downsampled_channel_0.tiff",
+ # "downsampled_standard.tiff",
+ # "downsampled_standard_channel_0.tiff",
+ # "registered_atlas.tiff",
+ # "registered_hemispheres.tiff",
+ ]
+ output_directory = os.path.join(output_directory, "registration")
+ for image in image_list:
+ are_images_equal(image, output_directory, test_niftyreg_output)
+
+ if platform.system() == "Linux":
+ pd.testing.assert_frame_equal(
+ pd.read_csv(os.path.join(output_directory, "volumes.csv")),
+ pd.read_csv(os.path.join(test_niftyreg_output, "volumes.csv")),
+ )
+
+
+def are_images_equal(image_name, output_directory, test_output_directory):
+ image = load_any(
+ os.path.join(output_directory, image_name),
+ )
+ test_image = load_any(
+ os.path.join(test_output_directory, image_name),
+ )
+ np.testing.assert_allclose(
+ image, test_image, rtol=relative_tolerance, atol=absolute_tolerance
+ )
diff --git a/tests/tests/test_unit/test_tools/test_image_processing.py b/tests/tests/test_unit/test_tools/test_image_processing.py
new file mode 100644
index 00000000..788fa01d
--- /dev/null
+++ b/tests/tests/test_unit/test_tools/test_image_processing.py
@@ -0,0 +1,37 @@
+import random
+
+import numpy as np
+
+from cellfinder.tools import image_processing as img_tools
+
+
+def test_crop_center_2d():
+ x_shape = random.randint(2, 100)
+ y_shape = random.randint(2, 100)
+ img = np.random.rand(y_shape, x_shape)
+ assert (
+ img == img_tools.crop_center_2d(img, crop_x=x_shape, crop_y=y_shape)
+ ).all()
+
+ new_x_shape = random.randint(1, x_shape)
+ new_y_shape = random.randint(1, y_shape)
+ pad_img = img_tools.crop_center_2d(
+ img, crop_x=new_x_shape, crop_y=new_y_shape
+ )
+ assert (new_y_shape, new_x_shape) == pad_img.shape
+
+
+def test_pad_centre_2d():
+ x_shape = random.randint(2, 100)
+ y_shape = random.randint(2, 100)
+ img = np.random.rand(y_shape, x_shape)
+ assert (
+ img == img_tools.pad_center_2d(img, x_size=x_shape, y_size=y_shape)
+ ).all()
+
+ new_x_shape = random.randint(x_shape, x_shape * 10)
+ new_y_shape = random.randint(y_shape, y_shape * 10)
+ pad_img = img_tools.pad_center_2d(
+ img, x_size=new_x_shape, y_size=new_y_shape
+ )
+ assert (new_y_shape, new_x_shape) == pad_img.shape
diff --git a/tests/tests/test_unit/test_tools/test_prep.py b/tests/tests/test_unit/test_tools/test_prep.py
new file mode 100644
index 00000000..404b5703
--- /dev/null
+++ b/tests/tests/test_unit/test_tools/test_prep.py
@@ -0,0 +1,241 @@
+import os
+
+import pytest
+from brainglobe_utils.general.exceptions import CommandLineInputError
+
+from cellfinder.tools import prep
+
+# import shutil
+
+# from pathlib import Path
+
+
+data_dir = os.path.join("tests", "data")
+
+
+def test_check_return_ch_ids():
+ signal_ch = [0, 1, 3]
+ bg_ch = 6
+ signal_list = ["file1.txt", "file_2.txt", "file_3.txt"]
+ # None given
+ assert ([0, 1, 2], 3) == prep.check_and_return_ch_ids(
+ None, None, signal_list
+ )
+ # Only signal given
+ assert (signal_ch, 4) == prep.check_and_return_ch_ids(
+ signal_ch, None, signal_list
+ )
+
+ # Only background given
+ assert ([7, 8, 9], bg_ch) == prep.check_and_return_ch_ids(
+ None, bg_ch, signal_list
+ )
+
+ # Both given (no overlap)
+ assert (signal_ch, bg_ch) == prep.check_and_return_ch_ids(
+ signal_ch, bg_ch, signal_list
+ )
+
+ # Both given (overlap)
+ with pytest.raises(CommandLineInputError):
+ assert prep.check_and_return_ch_ids(signal_ch, 3, signal_list)
+
+
+class Args:
+ def __init__(
+ self,
+ model_dir=None,
+ empty=None,
+ no_detection=False,
+ no_classification=False,
+ no_register=False,
+ no_analyse=False,
+ no_standard_space=False,
+ output_dir=None,
+ registration_output_folder=None,
+ cells_file_path=None,
+ cubes_output_dir=None,
+ classification_out_file=None,
+ cells_in_standard_space=None,
+ ):
+ self.cell_count_model_dir = model_dir
+ self.empty = empty
+
+ self.no_detection = no_detection
+ self.no_classification = no_classification
+ self.no_register = no_register
+ self.no_summarise = no_analyse
+ self.no_standard_space = no_standard_space
+
+ self.output_dir = output_dir
+
+ self.paths = Paths(
+ output_dir=registration_output_folder,
+ cells_file_path=cells_file_path,
+ cubes_output_dir=cubes_output_dir,
+ classification_out_file=classification_out_file,
+ cells_in_standard_space=cells_in_standard_space,
+ )
+
+
+class Paths:
+ def __init__(
+ self,
+ output_dir=None,
+ cells_file_path=None,
+ cubes_output_dir=None,
+ classification_out_file=None,
+ cells_in_standard_space=None,
+ ):
+ self.registration_output_folder = output_dir
+ self.cells_file_path = cells_file_path
+ self.tmp__cubes_output_dir = cubes_output_dir
+ self.classification_out_file = classification_out_file
+ self.cells_in_standard_space = cells_in_standard_space
+
+
+# def get_dict_of_what_to_run(what_to_run):
+# what_to_run_dict = {
+# "detect": what_to_run.detect,
+# "classify": what_to_run.classify,
+# "register": what_to_run.register,
+# "figures": what_to_run.figures,
+# }
+# return what_to_run_dict
+#
+#
+# def test_calc_what_to_run(tmpdir):
+# tmpdir = str(tmpdir)
+#
+# # registered_atlas_path = os.path.join(tmpdir, "registered_atlas.tiff")
+# cells_file_path = os.path.join(tmpdir, "cells.xml")
+# classification_out_file_path = os.path.join(
+# tmpdir, "cell_classification.xml"
+# )
+# summary_cell_counts_path = os.path.join(
+# tmpdir, "summary_cell_counts.csv")
+# figures_dir = os.path.join(tmpdir, "figures")
+#
+# args = Args(
+# output_dir=tmpdir,
+# registration_output_folder=tmpdir,
+# cells_file_path=cells_file_path,
+# classification_out_file=classification_out_file_path,
+# figures_dir=figures_dir,
+# )
+#
+# # default
+# what_to_run = prep.CalcWhatToRun(args)
+# default_validation = {
+# "detect": True,
+# "classify": True,
+# "register": False,
+# "figures": False,
+# }
+# default_test = get_dict_of_what_to_run(what_to_run)
+# assert default_validation == default_test
+#
+# # options
+# args.no_classification = True
+# what_to_run = prep.CalcWhatToRun(args)
+# validation = {
+# "detect": True,
+# "classify": False,
+# "register": False,
+# "figures": False,
+# }
+# test = get_dict_of_what_to_run(what_to_run)
+# assert validation == test
+#
+#
+# # # existance
+# # Path(registered_atlas_path).touch()
+# # what_to_run = prep.CalcWhatToRun(args)
+# # validation = {
+# # "detect": True,
+# # "classify": True,
+# # "register": False,
+# # "figures": True,
+# # }
+# # test = get_dict_of_what_to_run(what_to_run)
+# # assert validation == test
+#
+# Path(cells_file_path).touch()
+# what_to_run.update(args)
+# validation = {
+# "detect": False,
+# "classify": True,
+# "register": False,
+# "figures": True,
+# }
+# test = get_dict_of_what_to_run(what_to_run)
+# assert validation == test
+#
+# os.remove(cells_file_path)
+# what_to_run.update(args)
+# validation = {
+# "detect": True,
+# "classify": True,
+# "register": False,
+# "figures": True,
+# }
+# test = get_dict_of_what_to_run(what_to_run)
+# assert validation == test
+#
+# Path(classification_out_file_path).touch()
+# what_to_run.update(args)
+# validation = {
+# "detect": False,
+# "classify": False,
+# "register": False,
+# "figures": True,
+# }
+# test = get_dict_of_what_to_run(what_to_run)
+# assert validation == test
+#
+# Path(summary_cell_counts_path).touch()
+# what_to_run.update(args)
+# validation = {
+# "detect": False,
+# "classify": False,
+# "register": False,
+# "summarise": False,
+# "figures": True,
+# }
+# test = get_dict_of_what_to_run(what_to_run)
+# assert validation == test
+#
+# make_and_fill_directory(figures_dir)
+# args.figures = True
+# what_to_run.update(args)
+# validation = {
+# "detect": False,
+# "classify": False,
+# "register": False,
+# "summarise": False,
+# "figures": False,
+# }
+# test = get_dict_of_what_to_run(what_to_run)
+# assert validation == test
+#
+#
+# # os.remove(registered_atlas_path)
+# os.remove(summary_cell_counts_path)
+# shutil.rmtree(figures_dir)
+# what_to_run.update(args)
+# validation = {
+# "detect": False,
+# "classify": False,
+# "register": True,
+# "summarise": True,
+# "figures": True,
+# }
+# test = get_dict_of_what_to_run(what_to_run)
+# assert validation == test
+
+
+def make_and_fill_directory(directory):
+ os.mkdir(directory)
+ for file_size in range(100, 200, 20):
+ with open(os.path.join(directory, str(file_size)), "wb") as fout:
+ fout.write(os.urandom(file_size))
diff --git a/tests/tests/test_unit/test_tools/test_system.py b/tests/tests/test_unit/test_tools/test_system.py
new file mode 100644
index 00000000..480036d1
--- /dev/null
+++ b/tests/tests/test_unit/test_tools/test_system.py
@@ -0,0 +1,99 @@
+import os
+from math import isclose
+from pathlib import Path
+
+import pytest
+from brainglobe_utils.general.exceptions import CommandLineInputError
+from brainglobe_utils.general.system import ensure_directory_exists
+
+import cellfinder.tools.system as system
+
+data_dir = Path("tests", "data")
+background_im_dir = os.path.join(data_dir, "background")
+
+
+def test_get_subdirectories():
+ subdirs = system.get_subdirectories(data_dir)
+ assert len(subdirs) == 9
+ assert Path(data_dir / "cells") in subdirs
+ assert Path(data_dir / "brain") in subdirs
+
+ subdir_names = system.get_subdirectories(data_dir, names_only=True)
+ assert len(subdir_names) == 9
+ assert "cells" in subdir_names
+ assert "brain" in subdir_names
+
+
+def test_get_number_of_files_in_dir():
+ assert system.get_number_of_files_in_dir(background_im_dir) == 26
+
+
+def write_file_single_size(directory, file_size):
+ with open(os.path.join(directory, str(file_size)), "wb") as fout:
+ fout.write(os.urandom(file_size))
+
+
+def test_check_path_exists(tmpdir):
+ num = 10
+ tmpdir = str(tmpdir)
+
+ assert system.check_path_exists(os.path.join(tmpdir))
+ no_exist_dir = os.path.join(tmpdir, "i_dont_exist")
+ with pytest.raises(FileNotFoundError):
+ assert system.check_path_exists(no_exist_dir)
+
+ write_file_single_size(tmpdir, num)
+ assert system.check_path_exists(os.path.join(tmpdir, str(num)))
+ with pytest.raises(FileNotFoundError):
+ assert system.check_path_exists(os.path.join(tmpdir, "20"))
+
+
+def test_catch_input_file_error(tmpdir):
+ tmpdir = str(tmpdir)
+ # check no error is raised:
+ system.catch_input_file_error(tmpdir)
+
+ no_exist_dir = os.path.join(tmpdir, "i_dont_exist")
+ with pytest.raises(CommandLineInputError):
+ system.catch_input_file_error(no_exist_dir)
+
+
+def test_ensure_directory_exists():
+ home = os.path.expanduser("~")
+ exist_dir = os.path.join(home, ".cellfinder_test_dir")
+ ensure_directory_exists(exist_dir)
+ assert os.path.exists(exist_dir)
+ os.rmdir(exist_dir)
+
+
+def test_memory_in_bytes():
+ memory_detection_tolerance = 1 # byte
+
+ assert isclose(
+ system.memory_in_bytes(1, "kb"),
+ 1000,
+ abs_tol=memory_detection_tolerance,
+ )
+ assert isclose(
+ system.memory_in_bytes(1.2, "MB"),
+ 1200000,
+ abs_tol=memory_detection_tolerance,
+ )
+ assert isclose(
+ system.memory_in_bytes(0.00065, "gb"),
+ 650000,
+ abs_tol=memory_detection_tolerance,
+ )
+ assert isclose(
+ system.memory_in_bytes(0.000000000234, "TB"),
+ 234,
+ abs_tol=memory_detection_tolerance,
+ )
+ assert isclose(
+ system.memory_in_bytes(1000, "pb"),
+ 10**18,
+ abs_tol=memory_detection_tolerance,
+ )
+
+ with pytest.raises(NotImplementedError):
+ system.memory_in_bytes(1000, "ab")
diff --git a/tests/tests/test_unit/test_tools/test_tools_general.py b/tests/tests/test_unit/test_tools/test_tools_general.py
new file mode 100644
index 00000000..61cce44e
--- /dev/null
+++ b/tests/tests/test_unit/test_tools/test_tools_general.py
@@ -0,0 +1,65 @@
+import random
+
+import numpy as np
+import pytest
+
+import cellfinder.tools.tools as tools
+
+a = [1, "a", 10, 30]
+b = [30, 10, "c", "d"]
+
+test_2d_img = np.array([[1, 2, 10, 100], [5, 25, 300, 1000], [1, 0, 0, 125]])
+validate_2d_img = np.array(
+ [
+ [65.535, 131.07, 655.35, 6553.5],
+ [327.675, 1638.375, 19660.5, 65535],
+ [65.535, 0, 0, 8191.875],
+ ]
+)
+
+
+def test_get_max_value():
+ num = random.randint(0, 100)
+ assert 255 == tools.get_max_value(np.array(num, dtype=np.uint8))
+ assert 65535 == tools.get_max_value(np.array(num, dtype=np.uint16))
+
+
+def test_union():
+ ab = [1, "a", 10, 30, "c", "d"]
+ assert set(ab) == set(tools.union(a, b))
+
+
+def test_check_unique_list():
+ assert (True, []) == tools.check_unique_list(a)
+ repeating_list = [1, 2, 3, 3, "dog", "cat", "dog"]
+ assert (False, [3, "dog"]) == tools.check_unique_list(repeating_list)
+
+
+def test_common_member():
+ assert (True, [10, 30]) == tools.common_member(a, b)
+
+
+def test_get_number_of_bins_nd():
+ array_tuple = (100, 1000, 5000)
+ assert tools.get_number_of_bins_nd(array_tuple, 12) == (8, 83, 416)
+
+ shape_dict = {"x": 20, "y": 200}
+ assert tools.get_number_of_bins_nd(shape_dict, 8) == (2, 25)
+
+ with pytest.raises(NotImplementedError):
+ tools.get_number_of_bins_nd([200, 300, 400], 10)
+
+
+def test_interchange_np_fiji_coordinates():
+ tuple_in = (100, 200, 300)
+ assert tools.interchange_np_fiji_coordinates(tuple_in) == (200, 100, 300)
+
+
+def test_swap_elements_list():
+ list_in = [1, 2, 3, 4]
+ assert tools.swap_elements_list(list_in, 1, 2) == [1, 3, 2, 4]
+
+
+def test_is_any_list_overlap():
+ assert tools.is_any_list_overlap(a, b)
+ assert not tools.is_any_list_overlap(a, [2, "b", (1, 2, 3)])
diff --git a/tox.ini b/tox.ini
new file mode 100644
index 00000000..ed409e7a
--- /dev/null
+++ b/tox.ini
@@ -0,0 +1,25 @@
+# For more information about tox, see https://tox.readthedocs.io/en/latest/
+[tox]
+envlist = py{38,39,310}-{coredev}
+
+[gh-actions]
+python =
+ 3.8: py38
+ 3.9: py39
+ 3.10: py310
+
+[gh-actions:env]
+# This runs the coredev environment if the "coredev" github actions input
+# is set to "true"
+INPUT_COREDEV =
+ true: coredev
+
+[testenv]
+commands = pytest -v --cov=./ --cov-report=xml
+deps =
+ pytest-cov
+ pytest
+ coredev: git+https://github.com/brainglobe/cellfinder-core.git
+description =
+ Run tests
+ coredev: Run tests with the development version of cellfinder-core