diff --git a/mimic-iii-carevue/README.md b/mimic-iii-carevue/README.md
new file mode 100644
index 000000000..97484bd95
--- /dev/null
+++ b/mimic-iii-carevue/README.md
@@ -0,0 +1,129 @@
+# One-Click Installation Universal Script for MIMIC-III Clinical Database CareVue subset on MAC OS, Windows, and Linux
+
+[README](README.md) | [中文文档](README_zh.md)
+
+## 1 About this script
+
+A one-click installation universal script for MIMIC-III Clinical Database CareVue subset, no matter what operating system(MAC OS, Windows, and Linux), you can complete the installation of MIMIC-III Carevue database and related concepts in the fastest and most convenient way. A life-saving series tool for installation of critical care database.
+
+## 2 Before installation
+
+### 2.1 Install [Git](https://git-scm.com/download/win) (only for Windows user)
+
+In this script, `sed` would be used to configure **pg_hba.conf** (Reset the mode of local connection as `trust` with `sed` , for automatically and silently installation of the database with the shell script later). Mac OS terminals have ` sed`of `FreeBSD version` built in by default (of course you can also use `Homebrew` to install the `sed` of `GNU version` , namely `gsed`, not required), this script automatically recognizes the version of `sed` in MAC OS and executes the corresponding commands. The `sed` built in Linux distributions and [Git software](https://git-scm.com/download/win) for Windows is `gsed`. Since the shell scripts cannot be executed with `Command Prompt` on Windows (also unable to excute `sed` command), we would run shell scripts in `Terminal` provided by `Git`.
+
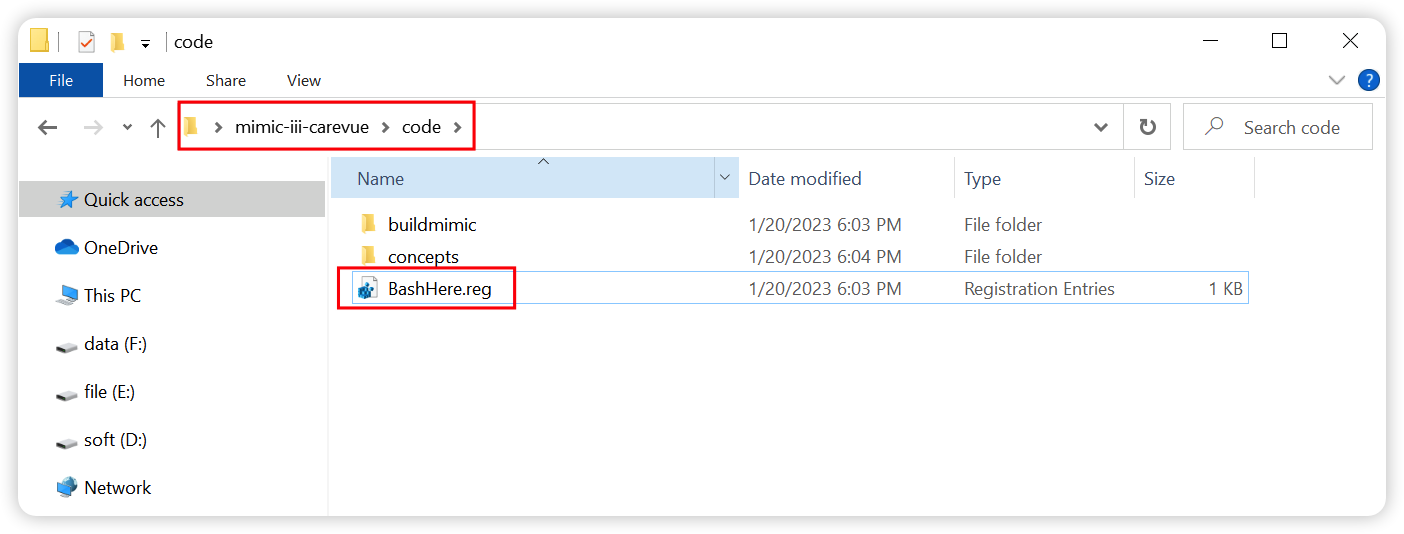
+#### ① After installation of [Git](https://git-scm.com/download/win), double click `BashHere.reg` to create shortcut of `bash Terminal`.
+
+Download the latest version of Git and install it with the default settings(**Install Git in `C:\Program Files\Git`**). Because the script will utilize the pipe operator for redirection, and terminal provided by `Git bash here` will cause the parameter passing failure of pipe operator, so we use the terminal provided by `bash.exe` in `C:\Program Files\Git\bin` to ensure successful execution of script. Next we would create shortcut of `bash Terminal ` to the right mouse menu by double-clicking `BashHere.reg` in the `code` subfolder.
+
+
+  +
+
+
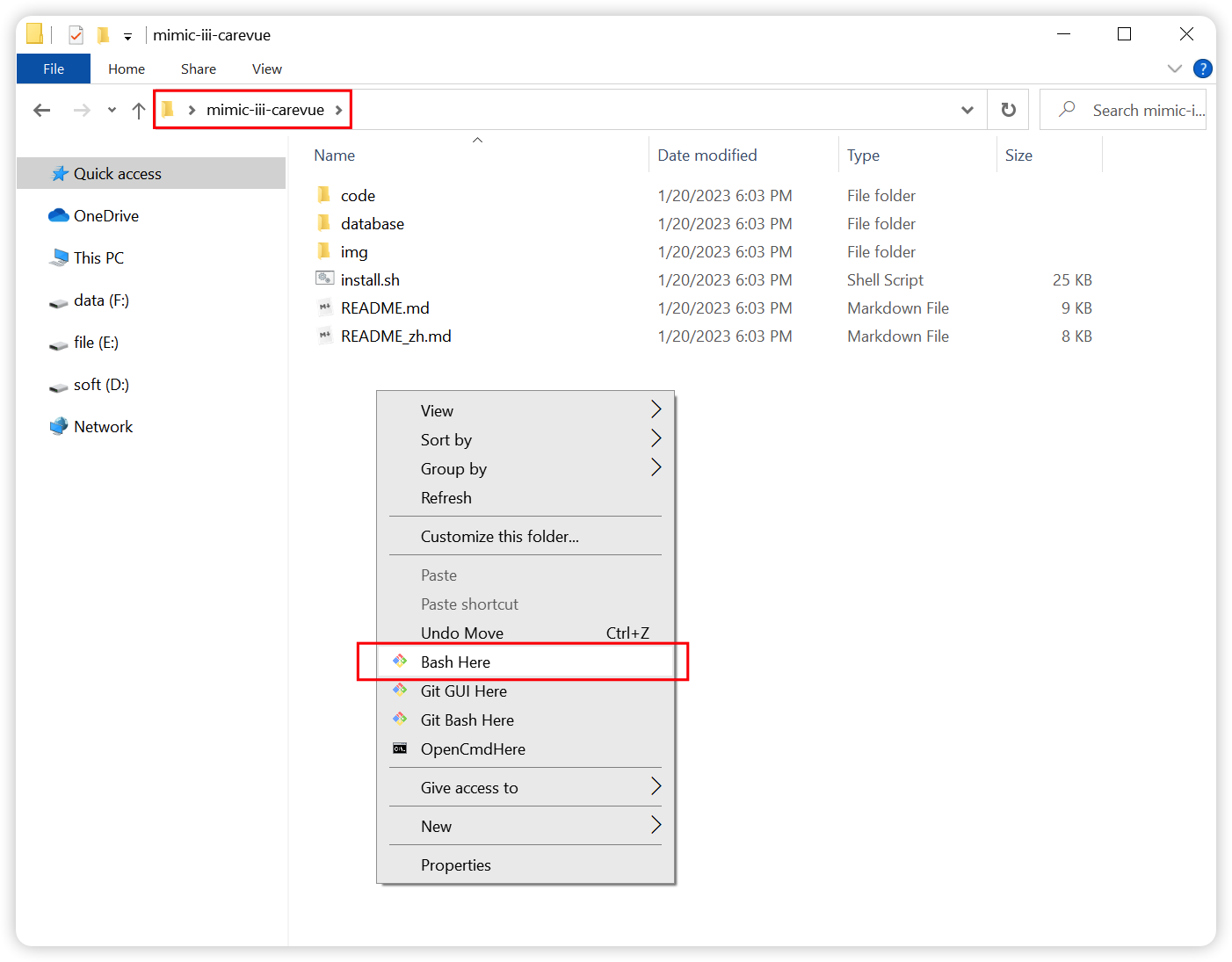
+#### ② Open the `bash Terminal`.
+
+Right-click in the `mimic-iii-carevue` folder and select `Bash Here` to open `bash Terminal`, which supports piping and redirection.
+
+
+  +
+
+
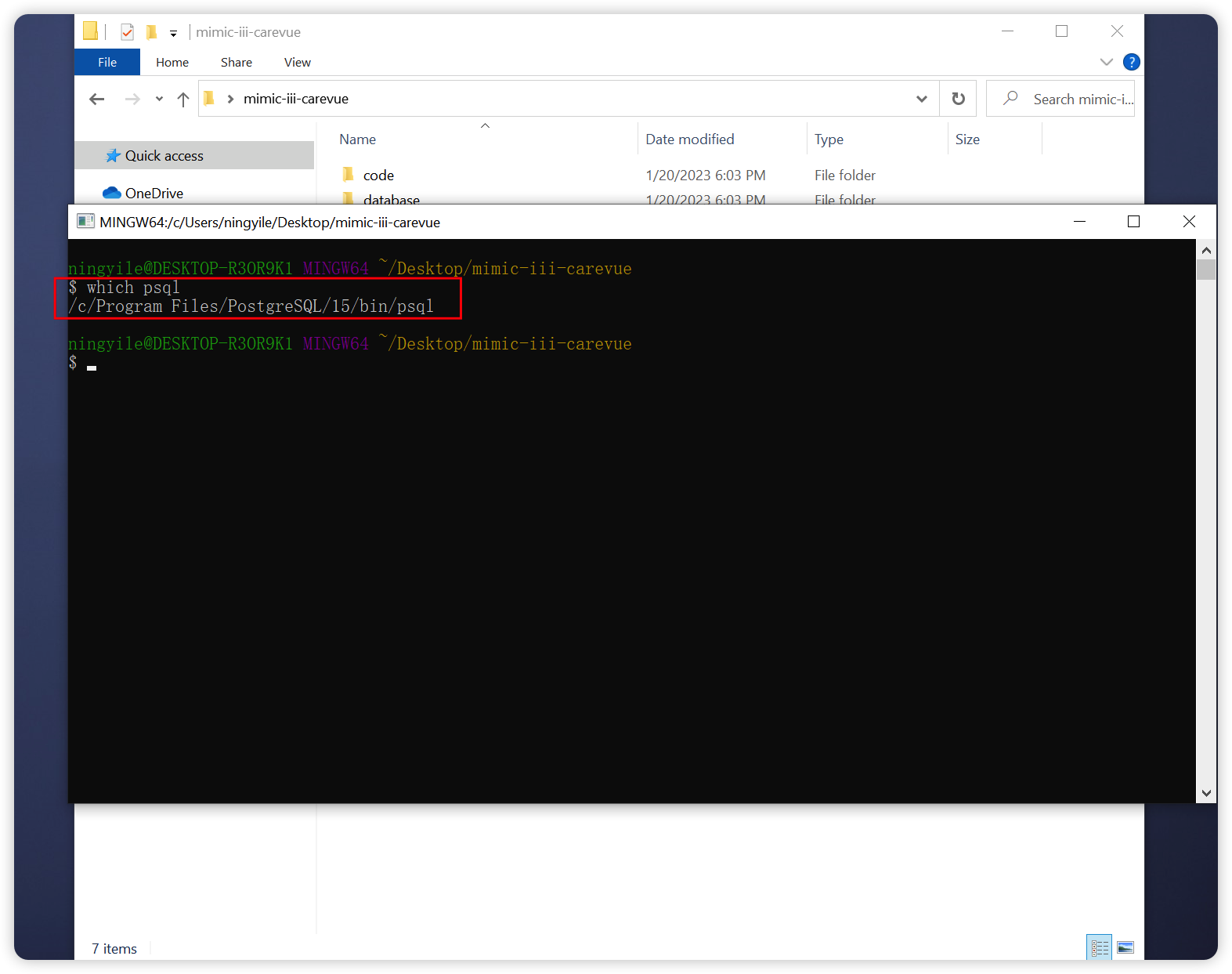
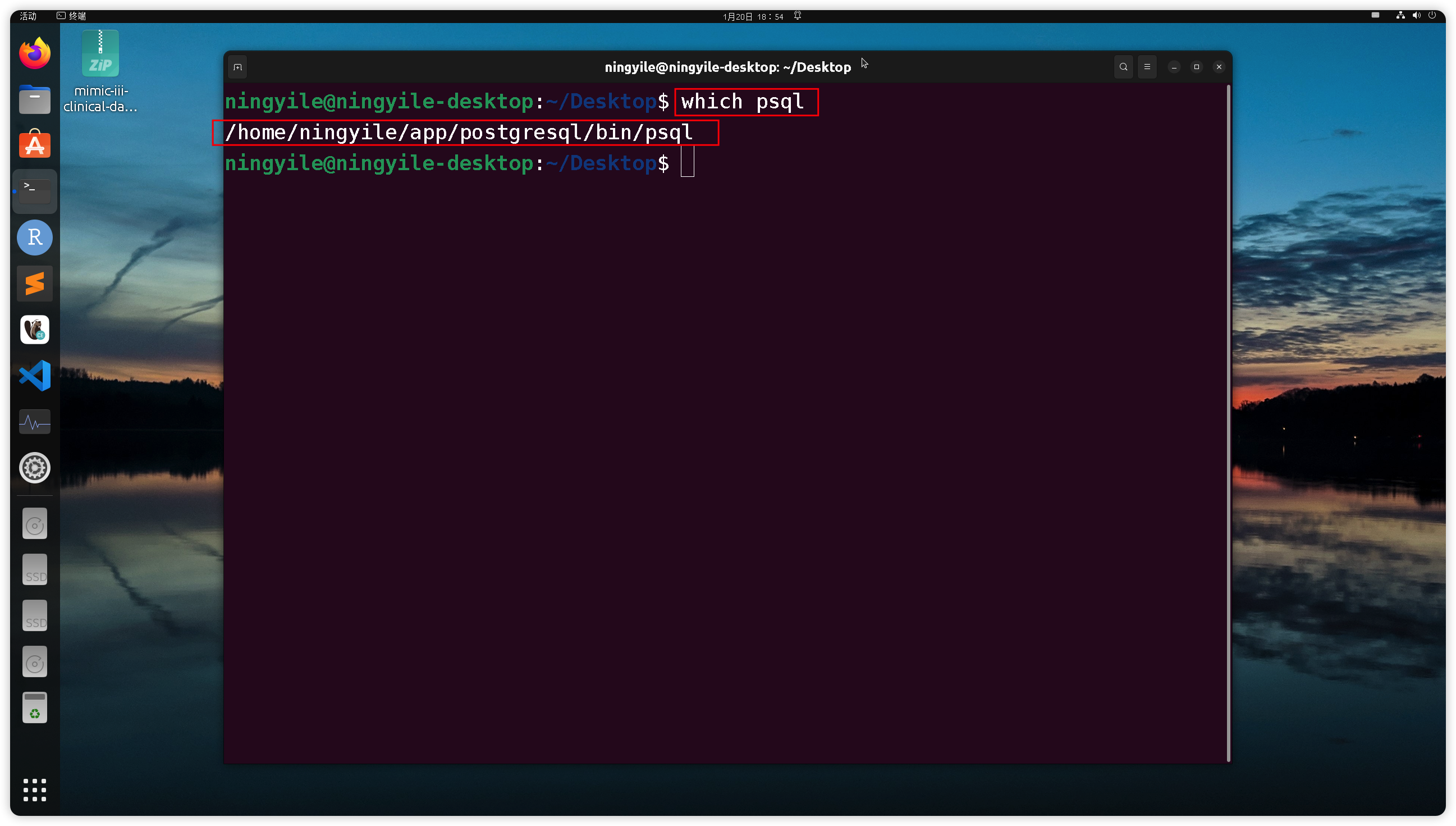
+### 2.2 Ensure Postgresql was installed and `psql` bin directory path was added to the environment variable (required)
+
+Open the `Terminal`(For Windows user, Right-click in the `mimic-iii-carevue` folder and select `Bash Here` to open `bash Terminal`, which mentioned in section 2.1). The environment variable was set correctly if a specific installation path (Postgresql ) is returned after type the following command in the `Terminal`.
+
+```shell
+which psql
+```
+- On Windows 10
+
+  +
+
+
+- On Ubuntu
+
+  +
+
+
+- On MAC OS
+
+ 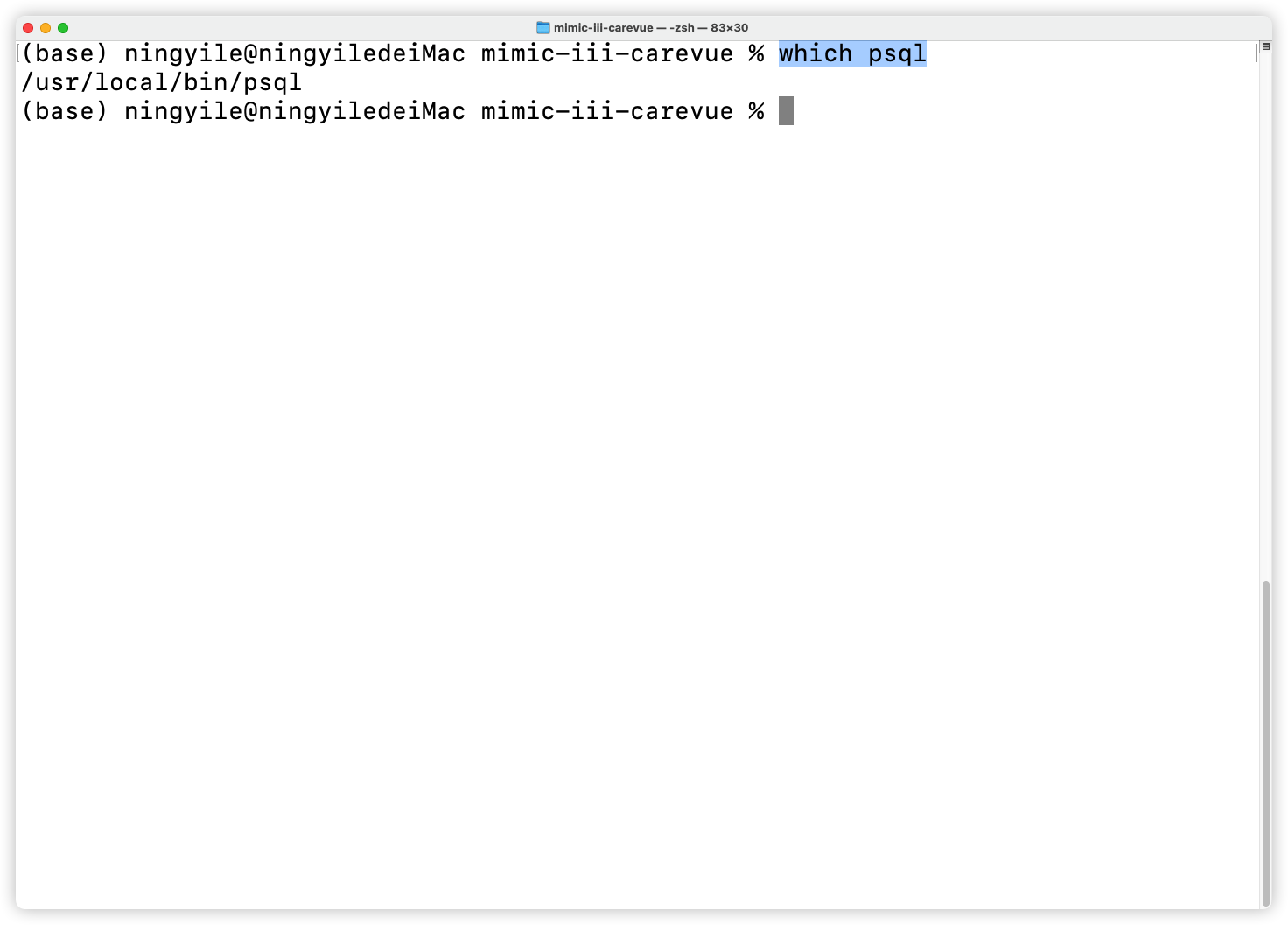 +
+
+
+### 2.3 Operating System User Name and Database User name (required)
+
+#### ① Ensure operating system user name does not contain any special characters and Spaces
+
+In order to facilitate the installation and maintenance of the database, the shell script will create the database user and database with the same name as the operating system user name. Then the `psql` command can be used directly in the terminal without any user name and database parameters (namely `-U` and `-d`). Therefore, the script first checks whether the operating system user name contains any special characters, such as Spaces, Chinese alphabets, Japanese alphabets, korean alphabets and so on. If the user name meet the preceding conditions, the installation will be automatically terminated unless creating a new user name that meets the conditions (preferably any combination of `English alphabets`, `numbers`, and`_`) and log in.
+
+#### ② Ensure the default database user name `postgres` is available
+After installing Postgresql with the default setting, the database will have an administrator user named `postgres`. The `psql` commands would be used in this one-click installation script, so make sure that the status of the database user `postgres` is available. Generally speaking, whether on MAC OS, Windows, or Ubuntu, the `postgres` administrator user status is available as long as the default settings were not changed. Run the following command in the terminal to check whether the `postgres`administrator user exists. If the terminal displays a message starting with `postgres`, it indicates that the `postgres` is available (The following figure for MAC OS as an example).
+
+```shell
+psql -U postgres;
+```
+
+ 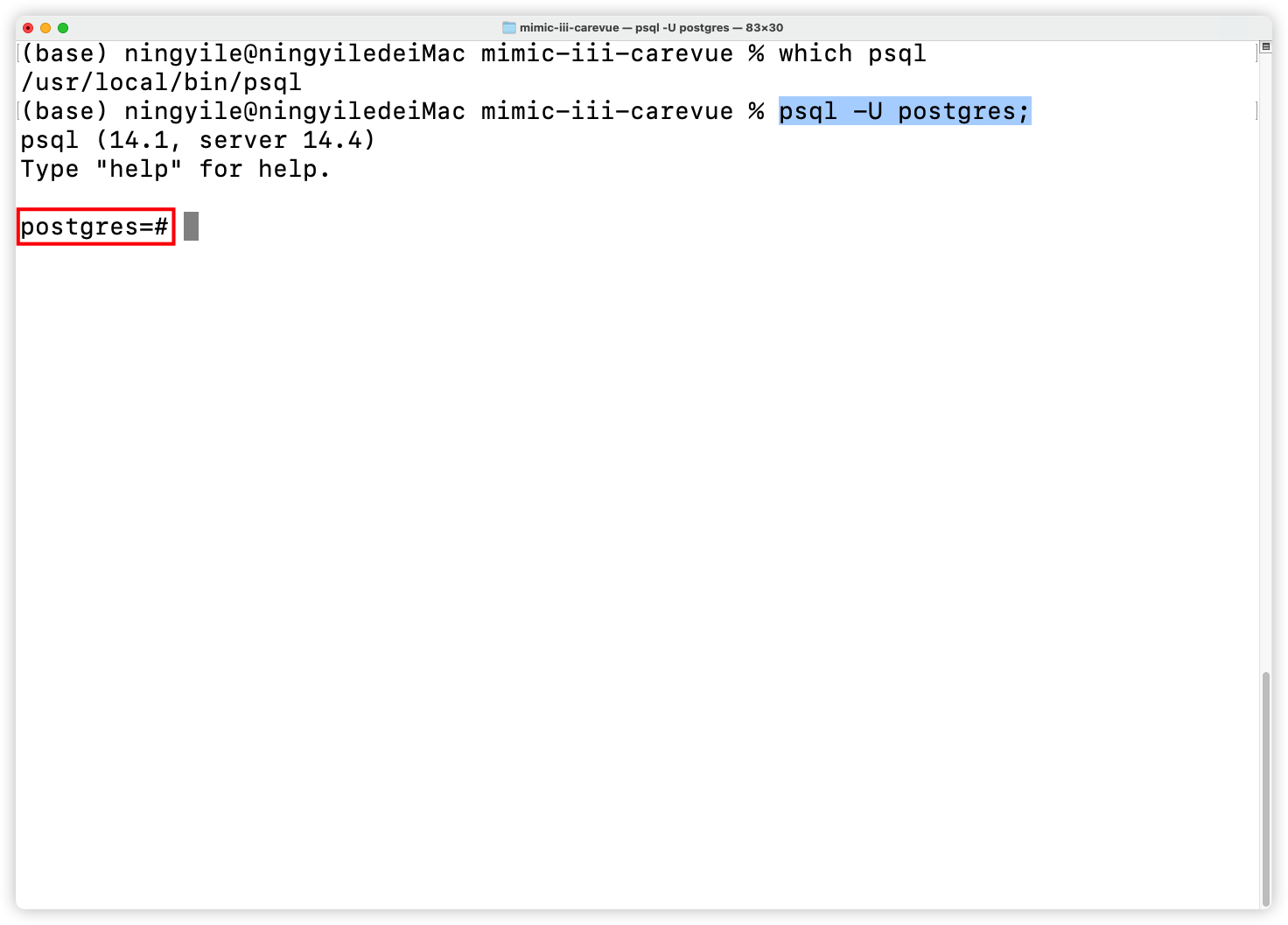 +
+
+
+
+## 3 Installation of MIMIC-III CareVue
+
+### 3.1 Load path of database files
+
+Decompress the `mimic-iii-clinical-database-carevue-subset-1.4.zip` database file and then move to the **database folder** (The following figure for MAC OS as an example).
+
+
+ 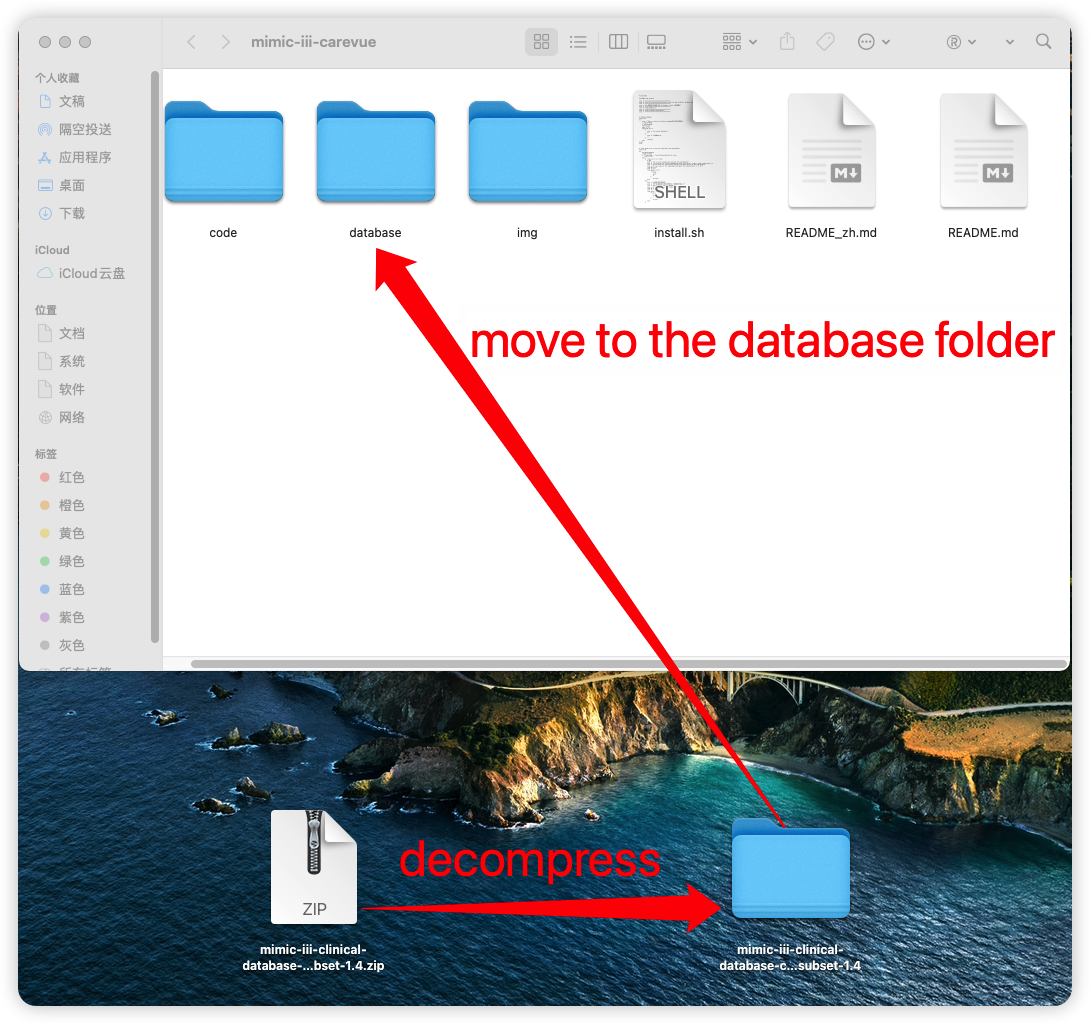 +
+
+### 3.2 One click installation in the `Terminal`
+
+Open the the `Terminal` (for Windows users, you need to right-click the mouse and choose `git bash here` to open the `Terminal` provided by `Git`) and enter **the path of install shell script**, type and run the following command, and select the corresponding option to complete the automatic installation of the MIMIC-III CareVue database.
+
+```shell
+bash install.sh
+```
+- Install database with one command
+
+ 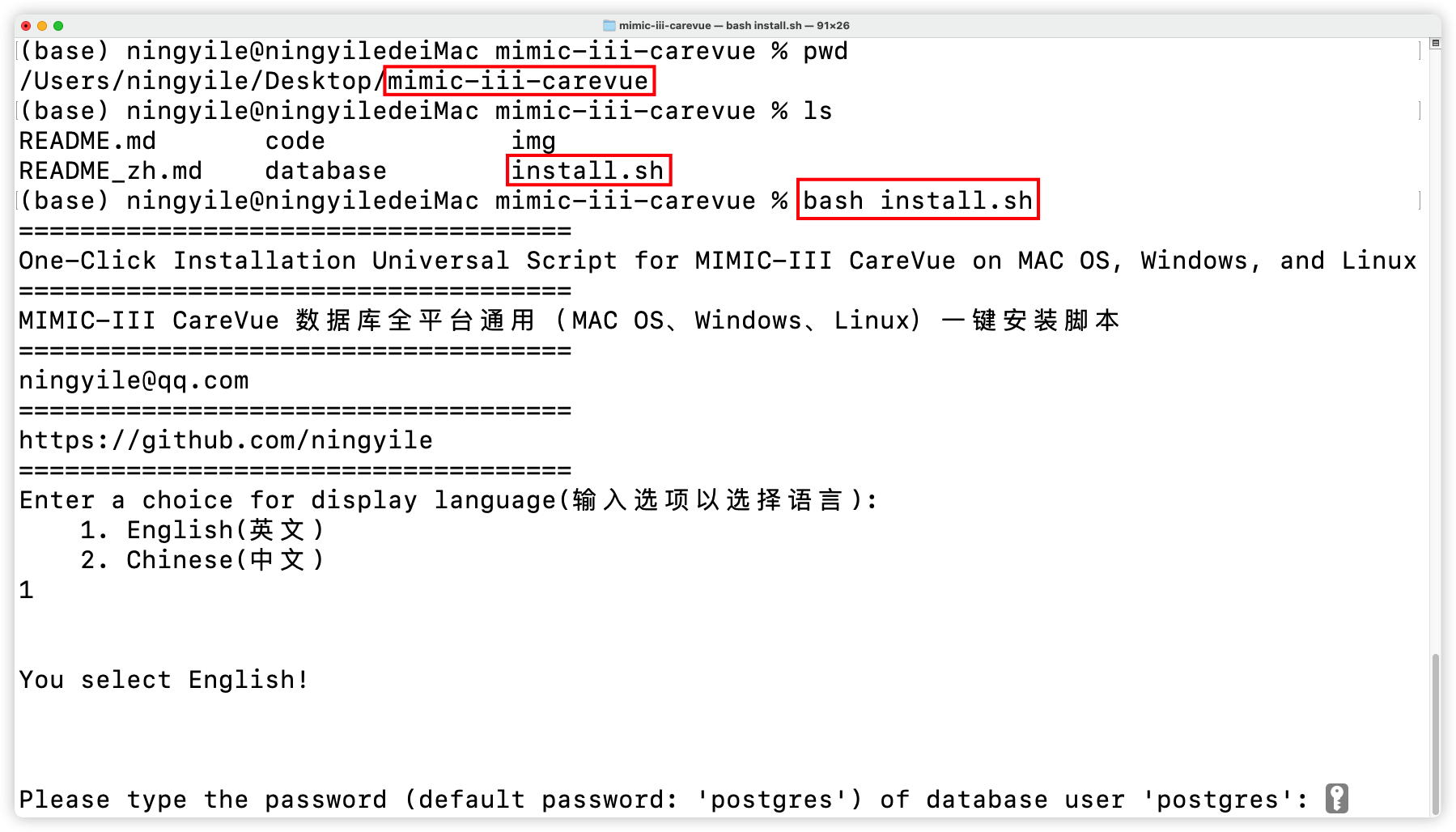 +
+
+
+- Install basic tables
+
+ 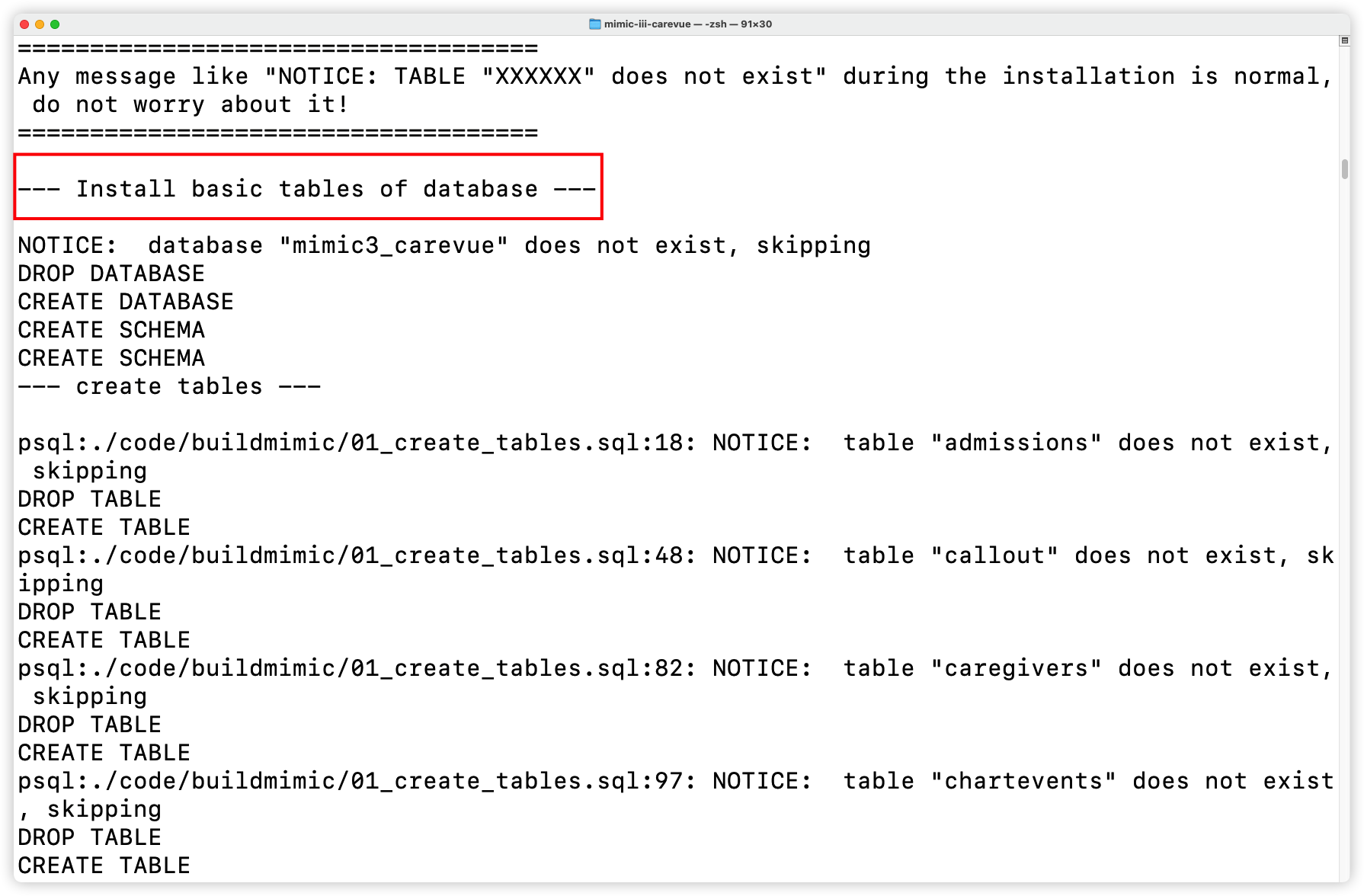 +
+
+
+- Install basic tables
+
+ 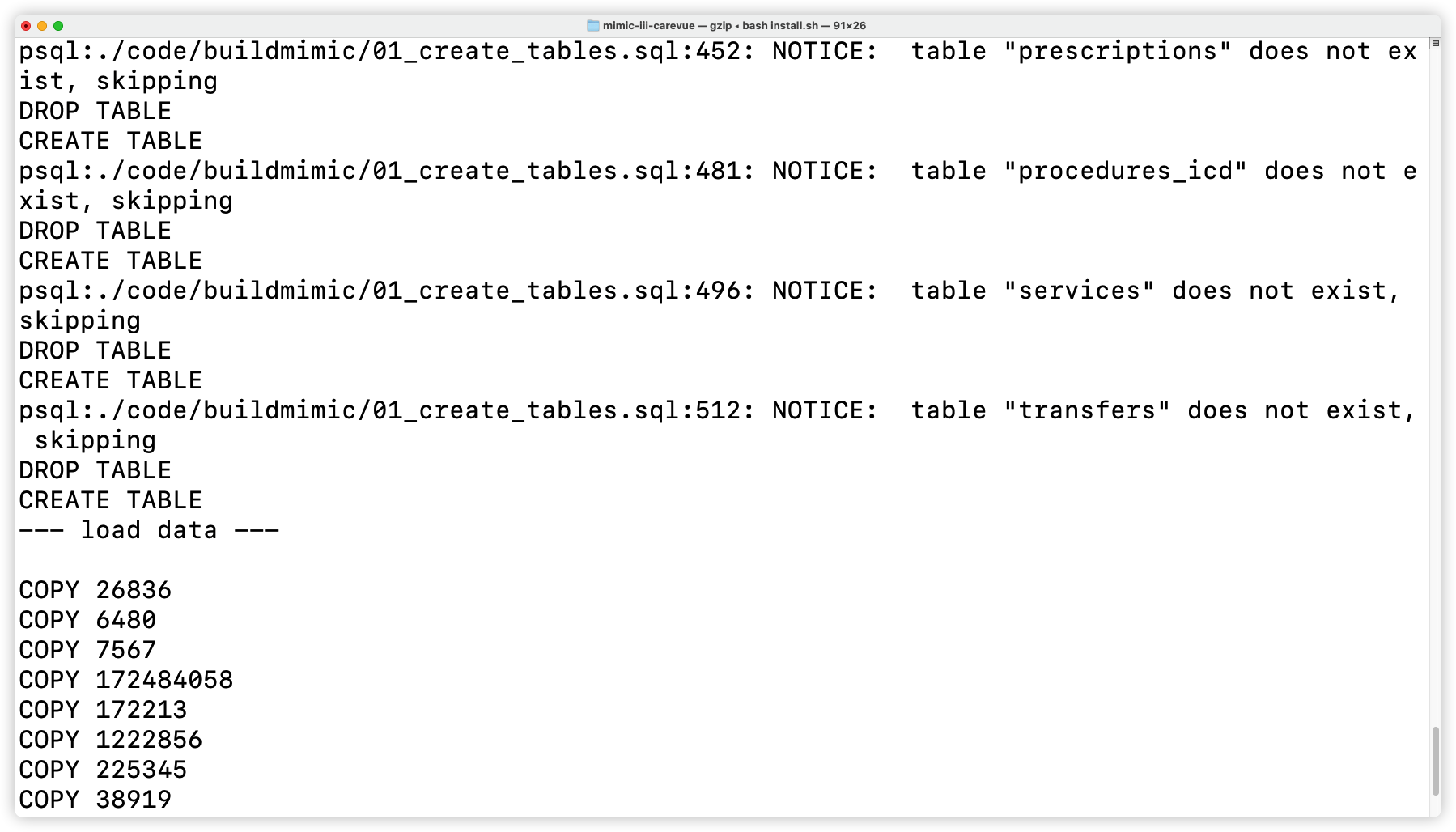 +
+
+
+- Install concepts
+
+ 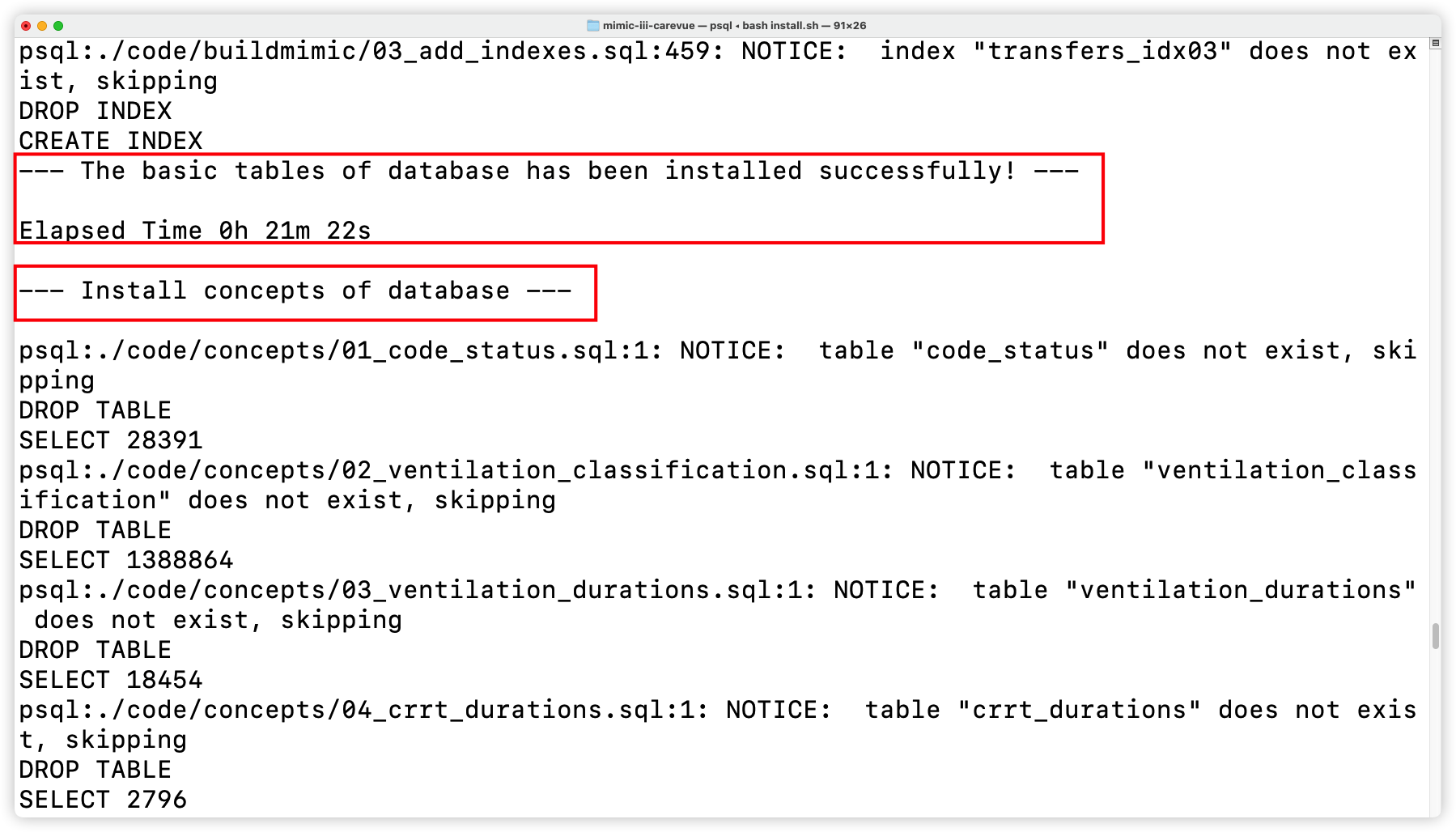 +
+
+
+- Check data
+
+ 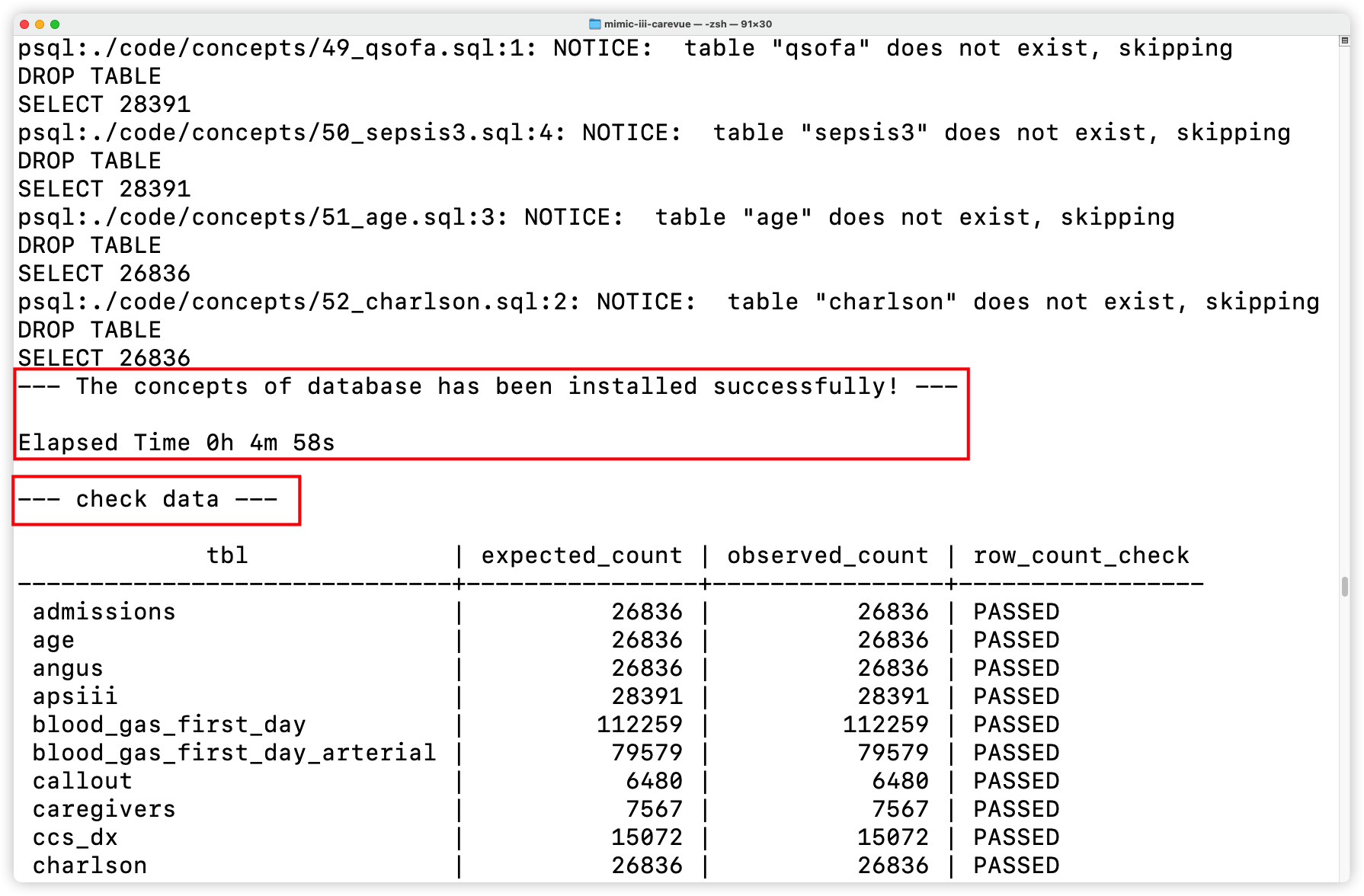 +
+
+
+- Check data
+
+ 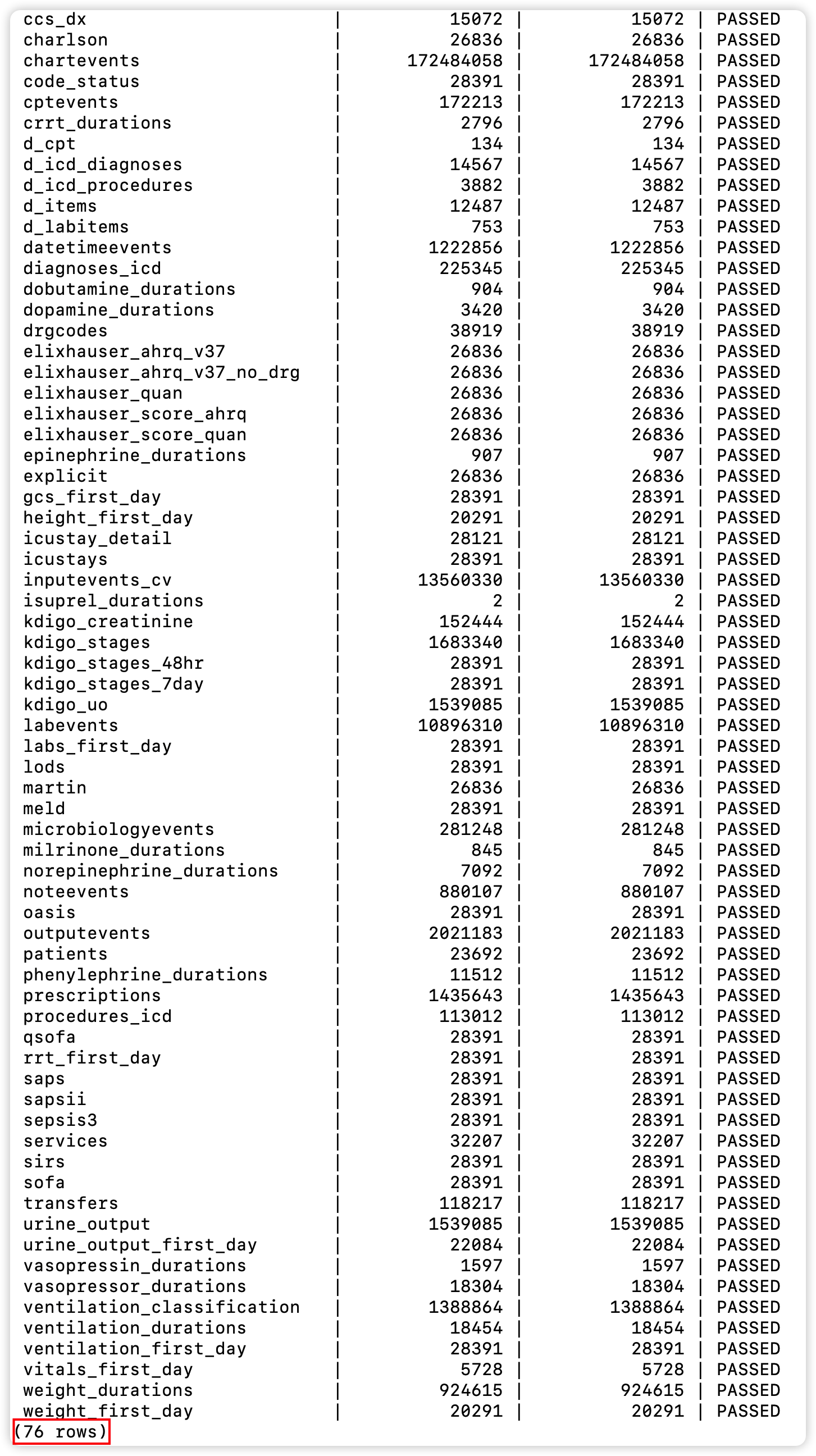 +
+
+
+- DBeaver
+
+ 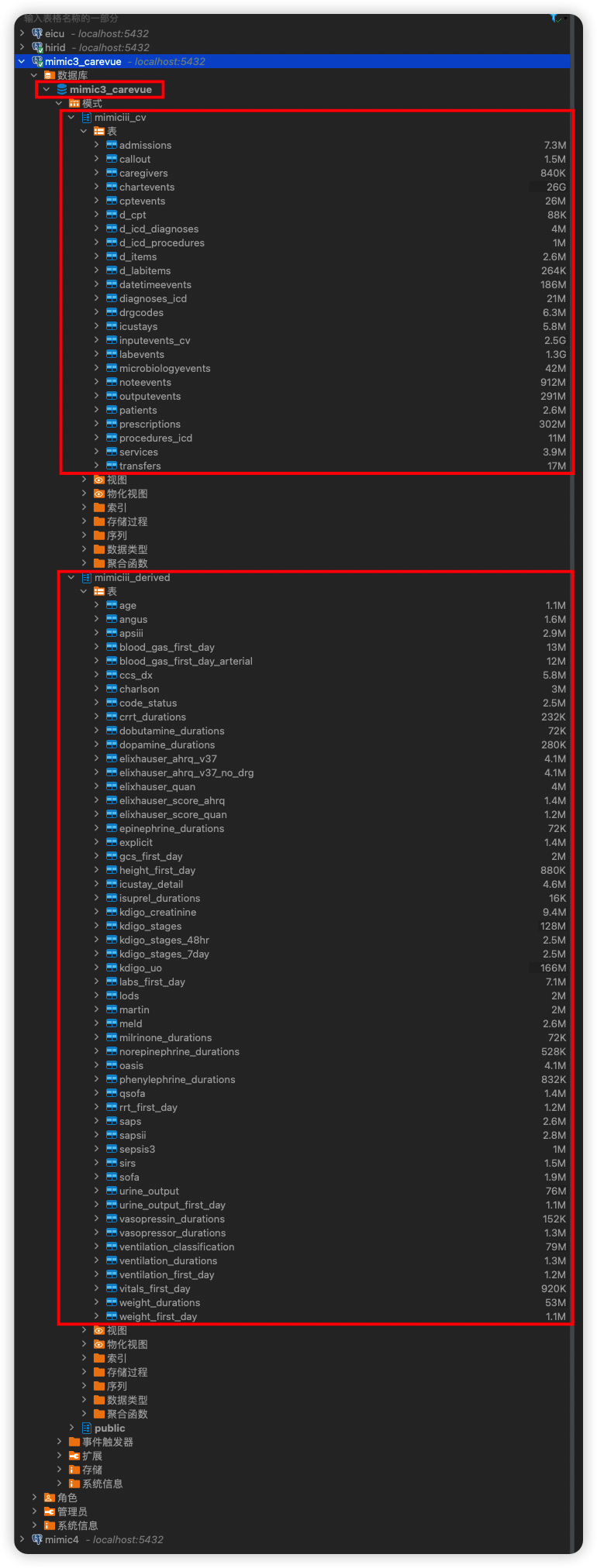 +
+
+
+## 4 P.S.
+
+* [MIMIC-III Clinical Database CareVue subset official website](https://physionet.org/content/mimic3-carevue/1.4/)
+* Related issue: [Are there duplications between patients in CareVue part of MIMIC III and patients in MIMIC IV ?](https://github.com/MIT-LCP/mimic-code/issues/1331)
+* SQL codes were edited based on the the base of [MIMICIII-V1.4](https://github.com/ningyile/mimic-code/tree/main/mimic-iii). Code blocks related to MetaVision (MV) were removed (including [echo_data.sql](https://github.com/ningyile/mimic-code/blob/main/mimic-iii/concepts/echo_data.sql)).
+* Sepsis3 has been added to the Concepts, which was edited based on [A Comparative Analysis of Sepsis Identification Methods in an Electronic Database](https://www.ncbi.nlm.nih.gov/pubmed/29303796). [sepsis3-mimic](https://github.com/alistairewj/sepsis3-mimic) created by[Alistair Johnson](https://github.com/alistairewj)。
+* Charlson Comorbidity Index has been added to the Concepts, which was edited based on[Charlson in Concepts of MIMICIV](https://github.com/MIT-LCP/mimic-code/blob/main/mimic-iv/concepts_postgres/comorbidity/charlson.sql)。
+* If you want to add a new feature, please create an issue first to describe the new feature.
+* Sorry for my poor English. Improvements for this document are welcome, even some typo fixes.
diff --git a/mimic-iii-carevue/README_zh.md b/mimic-iii-carevue/README_zh.md
new file mode 100644
index 000000000..277be816a
--- /dev/null
+++ b/mimic-iii-carevue/README_zh.md
@@ -0,0 +1,127 @@
+# MIMIC-III CareVue数据库全平台通用(MAC OS、Windows、Linux)一键安装脚本
+
+[README](README.md) | [中文文档](README_zh.md)
+
+## 1 关于此脚本
+
+一键式安装MIMIC-III CareVue数据库。无论什么操作系统(MAC OS、Windows、Linux),以最快、最方便的方式完成MIMIC-III CareVue数据库及相关Concepts的安装。此重症数据库系列安装脚本可节省你大量时间。
+
+## 2 安装前需要核对
+
+### 2.1 Windows用户请安装[Git软件](https://git-scm.com/download/win)(仅Windows用户必须)
+
+本脚本中需要使用`sed`命令**配置pg_hba.conf**(使用`sed`命令重置数据库的本地连接模式为`trust`,以便后期使用shell脚本自动静默安装数据库)。MAC OS终端默认`sed`版本为`FreeBSD`(当然你也可以使用`Homebrew`安装`GNU版本sed`,即`gsed`,非必须选项),此脚本可自动识别MAC OS终端`sed`版本并执行相应的命令。大多数Linux发行版以及Windows下[Git软件](https://git-scm.com/download/win)中内置的`sed`版本为`gsed`。由于Windows系统中`Command Prompt`没有`sed`命令且**无法运行shell脚本**,所以我们需要借用`Git终端`以确保脚本可以自动运行。
+
+#### ① 安装[Git软件](https://git-scm.com/download/win)后,双击 `BashHere.reg` 以创建 `bash`终端快捷键。
+
+下载最新版本的Git,然后使用默认设置进行安装(**确保安装到`C:\Program Files\Git`目录下**)。由于脚本中会使用到Unix的管道符的重定向进行操作,而Git软件默认的`Git bash here`会导致管道符传参失败,故我们使用`C:\Program Files\Git\bin`目录下的`bash.exe`作为终端确保脚本的顺利运行。接下来在`code`子文件夹中双击`BashHere.reg`添加`bash终端`的快捷键至鼠标右键。
+
+  +
+
+
+#### ② 右键打开 `bash`终端。
+
+在`mimic-iii-carevue文件夹`单击鼠标右键,选择`Bash Here`,即可打开支持管道符重定向的`bash终端`。
+
+  +
+
+
+### 2.2 确保Postgresql安装且环境变量已经配置(必须)
+
+打开 `终端`( **Windows用户单击鼠标右键,选择`Bash Here`**,使用2.1中提到的**`bash终端`**) ,输入以下代码,如返回一串具体的安装路径则证明`Postgresql环境变量`设置正常。
+
+```shell
+which psql
+```
+
+- On Windows 10
+
+  +
+
+
+- On Ubuntu
+
+  +
+
+
+- On MAC OS
+
+  +
+
+
+### 2.3 操作系统用户名及数据库用户名(必须)
+
+#### ① 确保操作系统用户名不要含有汉字、空格等特殊字符
+
+为方便日后数据库安装及维护,脚本会一键式设置与操作系统用户名同名的数据库用户及数据库,此后在终端可直接使用`psql`命令而无需附带任何用户名及数据库参数(即`-U`和`-d`)。因此脚本首先会判断操作系统用户名是否含有空格、汉字、日文、韩文等特殊字符,如符合上述条件,则安装将会自动终止,除非在操作系统下新建一个符合规范(尽量使用`英文字母`、`数字`以及`_`等字符任意组合)的用户名并以此登录。
+#### ② 确保默认的数据库用户`postgres`可用
+一般来说使用默认设置安装Postgresql之后,数据库会存在一个名为`postgres`的管理员用户。在这个一键安装脚本中会使用`终端`和`psql`命令进行一些操作,因此请确保`postgres管理员用户`的状态为可用。一般来说,无论在MAC OS、Windows或者Ubuntu,只要未修改默认设置,`postgres管理员用户`的状态都是可用的。在终端中输入以下命令即可检测`postgres管理员用户`是否存在(以MAC OS为例),当终端出现以`postgres`为开头的信息即表示可用。
+
+```shell
+psql -U postgres;
+```
+
+  +
+
+
+
+## 3 数据库一键安装
+
+### 3.1 数据库文件加载路径
+
+完成上述工作后,将`MIMIC3-Carevue-V1.4`数据库文件**解压后**放在**database文件夹**(以MAC OS为例)。
+
+  +
+
+### 3.2 一键安装
+
+在shell脚本文件路径下进入终端(对于Windows用户需要单击鼠标右键,选择`git bash here`,即可打开`Git终端`),输入并运行如下命令,选择相应的选项后即可完成数据库的自动安装。
+
+```shell
+bash install.sh
+```
+- 一键安装
+
+  +
+
+
+- 安装基础表单
+
+  +
+
+
+- 安装基础表单
+
+  +
+
+
+- 安装concepts
+
+  +
+
+
+- 数据核对
+
+  +
+
+
+- 数据核对
+
+  +
+
+
+- Dbeaver展示
+
+  +
+
+
+## 4 附言
+
+* [MIMIC-III Clinical Database CareVue subset official website](https://physionet.org/content/mimic3-carevue/1.4/)
+* 相关问题请参考:[Are there duplications between patients in CareVue part of MIMIC III and patients in MIMIC IV ?](https://github.com/MIT-LCP/mimic-code/issues/1331)
+* 安装代码修改自[MIMICIII-V1.4的sql代码](https://github.com/ningyile/mimic-code/tree/main/mimic-iii),移除了其中关于MetaVision(MV)相关数据源的安装代码(包括[echo_data.sql](https://github.com/ningyile/mimic-code/blob/main/mimic-iii/concepts/echo_data.sql))。
+* Concepts中增加了Sepsis3标准,其sql代码修改自[A Comparative Analysis of Sepsis Identification Methods in an Electronic Database](https://www.ncbi.nlm.nih.gov/pubmed/29303796)。[sepsis3-mimic](https://github.com/alistairewj/sepsis3-mimic) 由[Alistair Johnson](https://github.com/alistairewj)创立。
+* Concepts中增加了Charlson评分,其sql代码修改自[MIMICIV的Concepts中Charlson评分代码](https://github.com/MIT-LCP/mimic-code/blob/main/mimic-iv/concepts_postgres/comorbidity/charlson.sql)。
+* 如果您想添加一个新特性,请先创建一个问题来描述新特性。
+* 水平有限,欢迎对该文档进行改进,即使是一些拼写错误也可以。
+* 安装出现任何问题,欢迎通过ningyile@qq.com或者微信(`ningyile`)、推特(`ningyile`)联系(请备注来意)。
diff --git a/mimic-iii-carevue/code/BashHere.reg b/mimic-iii-carevue/code/BashHere.reg
new file mode 100644
index 000000000..a7820bd1e
--- /dev/null
+++ b/mimic-iii-carevue/code/BashHere.reg
@@ -0,0 +1,21 @@
+Windows Registry Editor Version 5.00
+
+[HKEY_CLASSES_ROOT\Directory\shell\Git Bash Here]
+
+"icon"="C:\\Program Files\\Git\\mingw64\\share\\git\\git-for-windows.ico"
+
+@="Bash Here"
+
+[HKEY_CLASSES_ROOT\Directory\shell\Bash Here\command]
+
+@="C:\\Program Files\\Git\\bin\\bash --login -i"
+
+[HKEY_CLASSES_ROOT\Directory\Background\shell\Bash Here]
+
+"icon"="C:\\Program Files\\Git\\mingw64\\share\\git\\git-for-windows.ico"
+
+@="Bash Here"
+
+[HKEY_CLASSES_ROOT\Directory\Background\shell\Bash Here\command]
+
+@="C:\\Program Files\\Git\\bin\\bash --login -i"
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/buildmimic/01_create_tables.sql b/mimic-iii-carevue/code/buildmimic/01_create_tables.sql
new file mode 100644
index 000000000..c11e6e0cb
--- /dev/null
+++ b/mimic-iii-carevue/code/buildmimic/01_create_tables.sql
@@ -0,0 +1,522 @@
+-- -------------------------------------------------------------------------------
+--
+-- Create basic tables of MIMIC-III CareVue Subset Database
+--
+-- -------------------------------------------------------------------------------
+
+--------------------------------------------------------
+-- DDL for Table ADMISSIONS
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS ADMISSIONS CASCADE;
+CREATE TABLE ADMISSIONS
+(
+ ROW_ID INT NOT NULL,
+ SUBJECT_ID INT NOT NULL,
+ HADM_ID INT NOT NULL,
+ ADMITTIME TIMESTAMP(0) NOT NULL,
+ DISCHTIME TIMESTAMP(0) NOT NULL,
+ DEATHTIME TIMESTAMP(0),
+ ADMISSION_TYPE VARCHAR(50) NOT NULL,
+ ADMISSION_LOCATION VARCHAR(50) NOT NULL,
+ DISCHARGE_LOCATION VARCHAR(50) NOT NULL,
+ INSURANCE VARCHAR(255) NOT NULL,
+ LANGUAGE VARCHAR(10),
+ RELIGION VARCHAR(50),
+ MARITAL_STATUS VARCHAR(50),
+ ETHNICITY VARCHAR(200) NOT NULL,
+ EDREGTIME TIMESTAMP(0),
+ EDOUTTIME TIMESTAMP(0),
+ DIAGNOSIS VARCHAR(255),
+ HOSPITAL_EXPIRE_FLAG SMALLINT,
+ HAS_CHARTEVENTS_DATA SMALLINT NOT NULL,
+ CONSTRAINT adm_rowid_pk PRIMARY KEY (ROW_ID),
+ CONSTRAINT adm_hadm_unique UNIQUE (HADM_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table CALLOUT
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS CALLOUT CASCADE;
+CREATE TABLE CALLOUT
+(
+ ROW_ID INT NOT NULL,
+ SUBJECT_ID INT NOT NULL,
+ HADM_ID INT NOT NULL,
+ SUBMIT_WARDID INT,
+ SUBMIT_CAREUNIT VARCHAR(15),
+ CURR_WARDID INT,
+ CURR_CAREUNIT VARCHAR(15),
+ CALLOUT_WARDID INT,
+ CALLOUT_SERVICE VARCHAR(10) NOT NULL,
+ REQUEST_TELE SMALLINT NOT NULL,
+ REQUEST_RESP SMALLINT NOT NULL,
+ REQUEST_CDIFF SMALLINT NOT NULL,
+ REQUEST_MRSA SMALLINT NOT NULL,
+ REQUEST_VRE SMALLINT NOT NULL,
+ CALLOUT_STATUS VARCHAR(20) NOT NULL,
+ CALLOUT_OUTCOME VARCHAR(20) NOT NULL,
+ DISCHARGE_WARDID INT,
+ ACKNOWLEDGE_STATUS VARCHAR(20) NOT NULL,
+ CREATETIME TIMESTAMP(0) NOT NULL,
+ UPDATETIME TIMESTAMP(0) NOT NULL,
+ ACKNOWLEDGETIME TIMESTAMP(0),
+ OUTCOMETIME TIMESTAMP(0) NOT NULL,
+ FIRSTRESERVATIONTIME TIMESTAMP(0),
+ CURRENTRESERVATIONTIME TIMESTAMP(0),
+ CONSTRAINT callout_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table CAREGIVERS
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS CAREGIVERS CASCADE;
+CREATE TABLE CAREGIVERS
+(
+ ROW_ID INT NOT NULL,
+ CGID INT NOT NULL,
+ LABEL VARCHAR(15),
+ DESCRIPTION VARCHAR(30),
+ CONSTRAINT cg_rowid_pk PRIMARY KEY (ROW_ID),
+ CONSTRAINT cg_cgid_unique UNIQUE (CGID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table CHARTEVENTS
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS CHARTEVENTS CASCADE;
+CREATE TABLE CHARTEVENTS
+(
+ ROW_ID INT NOT NULL,
+ SUBJECT_ID INT NOT NULL,
+ HADM_ID INT,
+ ICUSTAY_ID INT,
+ ITEMID INT,
+ CHARTTIME TIMESTAMP(0),
+ STORETIME TIMESTAMP(0),
+ CGID INT,
+ VALUE VARCHAR(255),
+ VALUENUM DOUBLE PRECISION,
+ VALUEUOM VARCHAR(50),
+ WARNING INT,
+ ERROR INT,
+ RESULTSTATUS VARCHAR(50),
+ STOPPED VARCHAR(50),
+ CONSTRAINT chartevents_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table CPTEVENTS
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS CPTEVENTS CASCADE;
+CREATE TABLE CPTEVENTS
+(
+ ROW_ID INT NOT NULL,
+ SUBJECT_ID INT NOT NULL,
+ HADM_ID INT NOT NULL,
+ COSTCENTER VARCHAR(10) NOT NULL,
+ CHARTDATE TIMESTAMP(0),
+ CPT_CD VARCHAR(10) NOT NULL,
+ CPT_NUMBER INT,
+ CPT_SUFFIX VARCHAR(5),
+ TICKET_ID_SEQ INT,
+ SECTIONHEADER VARCHAR(50),
+ SUBSECTIONHEADER VARCHAR(255),
+ DESCRIPTION VARCHAR(200),
+ CONSTRAINT cpt_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table DATETIMEEVENTS
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS DATETIMEEVENTS CASCADE;
+CREATE TABLE DATETIMEEVENTS
+(
+ ROW_ID INT NOT NULL,
+ SUBJECT_ID INT NOT NULL,
+ HADM_ID INT,
+ ICUSTAY_ID INT,
+ ITEMID INT NOT NULL,
+ CHARTTIME TIMESTAMP(0) NOT NULL,
+ STORETIME TIMESTAMP(0) NOT NULL,
+ CGID INT NOT NULL,
+ VALUE TIMESTAMP(0),
+ VALUEUOM VARCHAR(50) NOT NULL,
+ WARNING SMALLINT,
+ ERROR SMALLINT,
+ RESULTSTATUS VARCHAR(50),
+ STOPPED VARCHAR(50),
+ CONSTRAINT datetime_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table DIAGNOSES_ICD
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS DIAGNOSES_ICD CASCADE;
+CREATE TABLE DIAGNOSES_ICD
+(
+ ROW_ID INT NOT NULL,
+ SUBJECT_ID INT NOT NULL,
+ HADM_ID INT NOT NULL,
+ SEQ_NUM INT,
+ ICD9_CODE VARCHAR(10),
+ CONSTRAINT diagnosesicd_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table DRGCODES
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS DRGCODES CASCADE;
+CREATE TABLE DRGCODES
+(
+ ROW_ID INT NOT NULL,
+ SUBJECT_ID INT NOT NULL,
+ HADM_ID INT NOT NULL,
+ DRG_TYPE VARCHAR(20) NOT NULL,
+ DRG_CODE VARCHAR(20) NOT NULL,
+ DESCRIPTION VARCHAR(255),
+ DRG_SEVERITY SMALLINT,
+ DRG_MORTALITY SMALLINT,
+ CONSTRAINT drg_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table D_CPT
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS D_CPT CASCADE;
+CREATE TABLE D_CPT
+(
+ ROW_ID INT NOT NULL,
+ CATEGORY SMALLINT NOT NULL,
+ SECTIONRANGE VARCHAR(100) NOT NULL,
+ SECTIONHEADER VARCHAR(50) NOT NULL,
+ SUBSECTIONRANGE VARCHAR(100) NOT NULL,
+ SUBSECTIONHEADER VARCHAR(255) NOT NULL,
+ CODESUFFIX VARCHAR(5),
+ MINCODEINSUBSECTION INT NOT NULL,
+ MAXCODEINSUBSECTION INT NOT NULL,
+ CONSTRAINT dcpt_ssrange_unique UNIQUE (SUBSECTIONRANGE),
+ CONSTRAINT dcpt_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table D_ICD_DIAGNOSES
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS D_ICD_DIAGNOSES CASCADE;
+CREATE TABLE D_ICD_DIAGNOSES
+(
+ ROW_ID INT NOT NULL,
+ ICD9_CODE VARCHAR(10) NOT NULL,
+ SHORT_TITLE VARCHAR(50) NOT NULL,
+ LONG_TITLE VARCHAR(255) NOT NULL,
+ CONSTRAINT d_icd_diag_code_unique UNIQUE (ICD9_CODE),
+ CONSTRAINT d_icd_diag_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table D_ICD_PROCEDURES
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS D_ICD_PROCEDURES CASCADE;
+CREATE TABLE D_ICD_PROCEDURES
+(
+ ROW_ID INT NOT NULL,
+ ICD9_CODE VARCHAR(10) NOT NULL,
+ SHORT_TITLE VARCHAR(50) NOT NULL,
+ LONG_TITLE VARCHAR(255) NOT NULL,
+ CONSTRAINT d_icd_proc_code_unique UNIQUE (ICD9_CODE),
+ CONSTRAINT d_icd_proc_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table D_ITEMS
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS D_ITEMS CASCADE;
+CREATE TABLE D_ITEMS
+(
+ ROW_ID INT NOT NULL,
+ ITEMID INT NOT NULL,
+ LABEL VARCHAR(200),
+ ABBREVIATION VARCHAR(100),
+ DBSOURCE VARCHAR(20),
+ LINKSTO VARCHAR(50),
+ CATEGORY VARCHAR(100),
+ UNITNAME VARCHAR(100),
+ PARAM_TYPE VARCHAR(30),
+ CONCEPTID INT,
+ CONSTRAINT ditems_itemid_unique UNIQUE (ITEMID),
+ CONSTRAINT ditems_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table D_LABITEMS
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS D_LABITEMS CASCADE;
+CREATE TABLE D_LABITEMS
+(
+ ROW_ID INT NOT NULL,
+ ITEMID INT NOT NULL,
+ LABEL VARCHAR(100) NOT NULL,
+ FLUID VARCHAR(100) NOT NULL,
+ CATEGORY VARCHAR(100) NOT NULL,
+ LOINC_CODE VARCHAR(100),
+ CONSTRAINT dlabitems_itemid_unique UNIQUE (ITEMID),
+ CONSTRAINT dlabitems_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table ICUSTAYS
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS ICUSTAYS CASCADE;
+CREATE TABLE ICUSTAYS
+(
+ ROW_ID INT NOT NULL,
+ SUBJECT_ID INT NOT NULL,
+ HADM_ID INT NOT NULL,
+ ICUSTAY_ID INT NOT NULL,
+ DBSOURCE VARCHAR(20) NOT NULL,
+ FIRST_CAREUNIT VARCHAR(20) NOT NULL,
+ LAST_CAREUNIT VARCHAR(20) NOT NULL,
+ FIRST_WARDID SMALLINT NOT NULL,
+ LAST_WARDID SMALLINT NOT NULL,
+ INTIME TIMESTAMP(0) NOT NULL,
+ OUTTIME TIMESTAMP(0),
+ LOS DOUBLE PRECISION,
+ CONSTRAINT icustay_icustayid_unique UNIQUE (ICUSTAY_ID),
+ CONSTRAINT icustay_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table INPUTEVENTS_CV
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS INPUTEVENTS_CV CASCADE;
+CREATE TABLE INPUTEVENTS_CV
+(
+ ROW_ID INT NOT NULL,
+ SUBJECT_ID INT NOT NULL,
+ HADM_ID INT,
+ ICUSTAY_ID INT,
+ CHARTTIME TIMESTAMP(0),
+ ITEMID INT,
+ AMOUNT DOUBLE PRECISION,
+ AMOUNTUOM VARCHAR(30),
+ RATE DOUBLE PRECISION,
+ RATEUOM VARCHAR(30),
+ STORETIME TIMESTAMP(0),
+ CGID INT,
+ ORDERID INT,
+ LINKORDERID INT,
+ STOPPED VARCHAR(30),
+ NEWBOTTLE INT,
+ ORIGINALAMOUNT DOUBLE PRECISION,
+ ORIGINALAMOUNTUOM VARCHAR(30),
+ ORIGINALROUTE VARCHAR(30),
+ ORIGINALRATE DOUBLE PRECISION,
+ ORIGINALRATEUOM VARCHAR(30),
+ ORIGINALSITE VARCHAR(30),
+ CONSTRAINT inputevents_cv_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table LABEVENTS
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS LABEVENTS CASCADE;
+CREATE TABLE LABEVENTS
+(
+ ROW_ID INT NOT NULL,
+ SUBJECT_ID INT NOT NULL,
+ HADM_ID INT,
+ ITEMID INT NOT NULL,
+ CHARTTIME TIMESTAMP(0),
+ VALUE VARCHAR(200),
+ VALUENUM DOUBLE PRECISION,
+ VALUEUOM VARCHAR(20),
+ FLAG VARCHAR(20),
+ CONSTRAINT labevents_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table MICROBIOLOGYEVENTS
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS MICROBIOLOGYEVENTS CASCADE;
+CREATE TABLE MICROBIOLOGYEVENTS
+(
+ ROW_ID INT NOT NULL,
+ SUBJECT_ID INT NOT NULL,
+ HADM_ID INT,
+ CHARTDATE TIMESTAMP(0),
+ CHARTTIME TIMESTAMP(0),
+ SPEC_ITEMID INT,
+ SPEC_TYPE_DESC VARCHAR(100),
+ ORG_ITEMID INT,
+ ORG_NAME VARCHAR(100),
+ ISOLATE_NUM SMALLINT,
+ AB_ITEMID INT,
+ AB_NAME VARCHAR(30),
+ DILUTION_TEXT VARCHAR(10),
+ DILUTION_COMPARISON VARCHAR(20),
+ DILUTION_VALUE DOUBLE PRECISION,
+ INTERPRETATION VARCHAR(5),
+ CONSTRAINT micro_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table NOTEEVENTS
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS NOTEEVENTS CASCADE;
+CREATE TABLE NOTEEVENTS
+(
+ ROW_ID INT NOT NULL,
+ SUBJECT_ID INT NOT NULL,
+ HADM_ID INT,
+ CHARTDATE TIMESTAMP(0),
+ CHARTTIME TIMESTAMP(0),
+ STORETIME TIMESTAMP(0),
+ CATEGORY VARCHAR(50),
+ DESCRIPTION VARCHAR(255),
+ CGID INT,
+ ISERROR CHAR(1),
+ TEXT TEXT,
+ CONSTRAINT noteevents_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table OUTPUTEVENTS
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS OUTPUTEVENTS CASCADE;
+CREATE TABLE OUTPUTEVENTS
+(
+ ROW_ID INT NOT NULL,
+ SUBJECT_ID INT NOT NULL,
+ HADM_ID INT,
+ ICUSTAY_ID INT,
+ CHARTTIME TIMESTAMP(0),
+ ITEMID INT,
+ VALUE DOUBLE PRECISION,
+ VALUEUOM VARCHAR(30),
+ STORETIME TIMESTAMP(0),
+ CGID INT,
+ STOPPED VARCHAR(30),
+ NEWBOTTLE CHAR(1),
+ ISERROR INT,
+ CONSTRAINT outputevents_cv_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table PATIENTS
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS PATIENTS CASCADE;
+CREATE TABLE PATIENTS
+(
+ ROW_ID INT NOT NULL,
+ SUBJECT_ID INT NOT NULL,
+ GENDER VARCHAR(5) NOT NULL,
+ DOB TIMESTAMP(0) NOT NULL,
+ DOD TIMESTAMP(0),
+ DOD_HOSP TIMESTAMP(0),
+ DOD_SSN TIMESTAMP(0),
+ EXPIRE_FLAG INT NOT NULL,
+ CONSTRAINT pat_subid_unique UNIQUE (SUBJECT_ID),
+ CONSTRAINT pat_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table PRESCRIPTIONS
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS PRESCRIPTIONS CASCADE;
+CREATE TABLE PRESCRIPTIONS
+(
+ ROW_ID INT NOT NULL,
+ SUBJECT_ID INT NOT NULL,
+ HADM_ID INT NOT NULL,
+ ICUSTAY_ID INT,
+ STARTDATE TIMESTAMP(0),
+ ENDDATE TIMESTAMP(0),
+ DRUG_TYPE VARCHAR(100) NOT NULL,
+ DRUG VARCHAR(100) NOT NULL,
+ DRUG_NAME_POE VARCHAR(100),
+ DRUG_NAME_GENERIC VARCHAR(100),
+ FORMULARY_DRUG_CD VARCHAR(120),
+ GSN VARCHAR(200),
+ NDC VARCHAR(120),
+ PROD_STRENGTH VARCHAR(120),
+ DOSE_VAL_RX VARCHAR(120),
+ DOSE_UNIT_RX VARCHAR(120),
+ FORM_VAL_DISP VARCHAR(120),
+ FORM_UNIT_DISP VARCHAR(120),
+ ROUTE VARCHAR(120),
+ CONSTRAINT prescription_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table PROCEDURES_ICD
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS PROCEDURES_ICD CASCADE;
+CREATE TABLE PROCEDURES_ICD
+(
+ ROW_ID INT NOT NULL,
+ SUBJECT_ID INT NOT NULL,
+ HADM_ID INT NOT NULL,
+ SEQ_NUM INT NOT NULL,
+ ICD9_CODE VARCHAR(10) NOT NULL,
+ CONSTRAINT proceduresicd_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table SERVICES
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS SERVICES CASCADE;
+CREATE TABLE SERVICES
+(
+ ROW_ID INT NOT NULL,
+ SUBJECT_ID INT NOT NULL,
+ HADM_ID INT NOT NULL,
+ TRANSFERTIME TIMESTAMP(0) NOT NULL,
+ PREV_SERVICE VARCHAR(20),
+ CURR_SERVICE VARCHAR(20),
+ CONSTRAINT services_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
+
+--------------------------------------------------------
+-- DDL for Table TRANSFERS
+--------------------------------------------------------
+
+DROP TABLE IF EXISTS TRANSFERS CASCADE;
+CREATE TABLE TRANSFERS
+(
+ ROW_ID INT NOT NULL,
+ SUBJECT_ID INT NOT NULL,
+ HADM_ID INT NOT NULL,
+ ICUSTAY_ID INT,
+ DBSOURCE VARCHAR(20),
+ EVENTTYPE VARCHAR(20),
+ PREV_CAREUNIT VARCHAR(20),
+ CURR_CAREUNIT VARCHAR(20),
+ PREV_WARDID SMALLINT,
+ CURR_WARDID SMALLINT,
+ INTIME TIMESTAMP(0),
+ OUTTIME TIMESTAMP(0),
+ LOS DOUBLE PRECISION,
+ CONSTRAINT transfers_rowid_pk PRIMARY KEY (ROW_ID)
+) ;
diff --git a/mimic-iii-carevue/code/buildmimic/02_load_data_gz.sql b/mimic-iii-carevue/code/buildmimic/02_load_data_gz.sql
new file mode 100644
index 000000000..3caafe617
--- /dev/null
+++ b/mimic-iii-carevue/code/buildmimic/02_load_data_gz.sql
@@ -0,0 +1,153 @@
+-- -------------------------------------------------------------------------------
+--
+-- Load data into the MIMIC-III CareVue
+--
+-- -------------------------------------------------------------------------------
+
+-- Change to the directory containing the data files
+\cd :mimic_data_dir
+\cd ./mimic-iii-clinical-database-carevue-subset-1.4
+
+--------------------------------------------------------
+-- Load Data for Table ADMISSIONS
+--------------------------------------------------------
+
+\copy ADMISSIONS FROM PROGRAM 'gzip -dc ADMISSIONS.csv.gz' DELIMITER ',' CSV HEADER NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table CALLOUT
+--------------------------------------------------------
+
+\copy CALLOUT from PROGRAM 'gzip -dc CALLOUT.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table CAREGIVERS
+--------------------------------------------------------
+
+\copy CAREGIVERS from PROGRAM 'gzip -dc CAREGIVERS.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table CHARTEVENTS
+--------------------------------------------------------
+
+\copy CHARTEVENTS from PROGRAM 'gzip -dc CHARTEVENTS.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table CPTEVENTS
+--------------------------------------------------------
+
+\copy CPTEVENTS from PROGRAM 'gzip -dc CPTEVENTS.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table DATETIMEEVENTS
+--------------------------------------------------------
+
+\copy DATETIMEEVENTS from PROGRAM 'gzip -dc DATETIMEEVENTS.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table DIAGNOSES_ICD
+--------------------------------------------------------
+
+\copy DIAGNOSES_ICD from PROGRAM 'gzip -dc DIAGNOSES_ICD.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table DRGCODES
+--------------------------------------------------------
+
+\copy DRGCODES from PROGRAM 'gzip -dc DRGCODES.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table D_CPT
+--------------------------------------------------------
+
+\copy D_CPT from PROGRAM 'gzip -dc D_CPT.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table D_ICD_DIAGNOSES
+--------------------------------------------------------
+
+\copy D_ICD_DIAGNOSES from PROGRAM 'gzip -dc D_ICD_DIAGNOSES.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table D_ICD_PROCEDURES
+--------------------------------------------------------
+
+\copy D_ICD_PROCEDURES from PROGRAM 'gzip -dc D_ICD_PROCEDURES.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table D_ITEMS
+--------------------------------------------------------
+
+\copy D_ITEMS from PROGRAM 'gzip -dc D_ITEMS.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table D_LABITEMS
+--------------------------------------------------------
+
+\copy D_LABITEMS from PROGRAM 'gzip -dc D_LABITEMS.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table ICUSTAYS
+--------------------------------------------------------
+
+\copy ICUSTAYS from PROGRAM 'gzip -dc ICUSTAYS.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table INPUTEVENTS_CV
+--------------------------------------------------------
+
+\copy INPUTEVENTS_CV from PROGRAM 'gzip -dc INPUTEVENTS_CV.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table LABEVENTS
+--------------------------------------------------------
+
+\copy LABEVENTS from PROGRAM 'gzip -dc LABEVENTS.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table MICROBIOLOGYEVENTS
+--------------------------------------------------------
+
+\copy MICROBIOLOGYEVENTS from PROGRAM 'gzip -dc MICROBIOLOGYEVENTS.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table NOTEEVENTS
+--------------------------------------------------------
+
+\copy NOTEEVENTS from PROGRAM 'gzip -dc NOTEEVENTS.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table OUTPUTEVENTS
+--------------------------------------------------------
+
+\copy OUTPUTEVENTS from PROGRAM 'gzip -dc OUTPUTEVENTS.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table PATIENTS
+--------------------------------------------------------
+
+\copy PATIENTS from PROGRAM 'gzip -dc PATIENTS.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table PRESCRIPTIONS
+--------------------------------------------------------
+
+\copy PRESCRIPTIONS from PROGRAM 'gzip -dc PRESCRIPTIONS.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table PROCEDURES_ICD
+--------------------------------------------------------
+
+\copy PROCEDURES_ICD from PROGRAM 'gzip -dc PROCEDURES_ICD.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table SERVICES
+--------------------------------------------------------
+
+\copy SERVICES from PROGRAM 'gzip -dc SERVICES.csv.gz' delimiter ',' csv header NULL ''
+
+--------------------------------------------------------
+-- Load Data for Table TRANSFERS
+--------------------------------------------------------
+
+\copy TRANSFERS from PROGRAM 'gzip -dc TRANSFERS.csv.gz' delimiter ',' csv header NULL ''
diff --git a/mimic-iii-carevue/code/buildmimic/03_add_indexes.sql b/mimic-iii-carevue/code/buildmimic/03_add_indexes.sql
new file mode 100644
index 000000000..affa0cae2
--- /dev/null
+++ b/mimic-iii-carevue/code/buildmimic/03_add_indexes.sql
@@ -0,0 +1,469 @@
+-- ----------------------------------------------------------------
+--
+-- This is a script to add the MIMIC-III indexes for Postgres.
+--
+-- ----------------------------------------------------------------
+
+-- If running scripts individually, you can set the schema where all tables are created as follows:
+-- SET search_path TO mimiciii;
+
+-- Restoring the search path to its default value can be accomplished as follows:
+-- SET search_path TO "$user",public;
+
+-------------
+-- ADMISSIONS
+-------------
+
+DROP INDEX IF EXISTS ADMISSIONS_idx01;
+CREATE INDEX ADMISSIONS_idx01
+ ON ADMISSIONS (SUBJECT_ID);
+
+DROP INDEX IF EXISTS ADMISSIONS_idx02;
+CREATE INDEX ADMISSIONS_idx02
+ ON ADMISSIONS (HADM_ID);
+
+-- DROP INDEX IF EXISTS ADMISSIONS_idx03;
+-- CREATE INDEX ADMISSIONS_idx03
+-- ON ADMISSIONS (ADMISSION_TYPE);
+
+
+-----------
+--CALLOUT--
+-----------
+
+DROP INDEX IF EXISTS CALLOUT_idx01;
+CREATE INDEX CALLOUT_idx01
+ ON CALLOUT (SUBJECT_ID);
+
+DROP INDEX IF EXISTS CALLOUT_idx02;
+CREATE INDEX CALLOUT_idx02
+ ON CALLOUT (HADM_ID);
+
+-- DROP INDEX IF EXISTS CALLOUT_idx03;
+-- CREATE INDEX CALLOUT_idx03
+-- ON CALLOUT (CALLOUT_SERVICE);
+
+-- DROP INDEX IF EXISTS CALLOUT_idx04;
+-- CREATE INDEX CALLOUT_idx04
+-- ON CALLOUT (CURR_WARDID, CALLOUT_WARDID,
+-- DISCHARGE_WARDID);
+
+-- DROP INDEX IF EXISTS CALLOUT_idx05;
+-- CREATE INDEX CALLOUT_idx05
+-- ON CALLOUT (CALLOUT_STATUS,
+-- CALLOUT_OUTCOME);
+
+-- DROP INDEX IF EXISTS CALLOUT_idx06;
+-- CREATE INDEX CALLOUT_idx06
+-- ON CALLOUT (CREATETIME, UPDATETIME,
+-- ACKNOWLEDGETIME, OUTCOMETIME);
+
+---------------
+-- CAREGIVERS
+---------------
+
+-- DROP INDEX IF EXISTS CAREGIVERS_idx01;
+-- CREATE INDEX CAREGIVERS_idx01
+-- ON CAREGIVERS (CGID, LABEL);
+
+---------------
+-- CHARTEVENTS
+---------------
+
+DROP INDEX IF EXISTS CHARTEVENTS_idx01;
+CREATE INDEX CHARTEVENTS_idx01
+ ON CHARTEVENTS (CHARTTIME, STORETIME);
+
+DROP INDEX IF EXISTS CHARTEVENTS_idx02;
+CREATE INDEX CHARTEVENTS_idx02
+ ON CHARTEVENTS (SUBJECT_ID);
+
+DROP INDEX IF EXISTS CHARTEVENTS_idx03;
+CREATE INDEX CHARTEVENTS_idx03
+ ON CHARTEVENTS (HADM_ID);
+
+DROP INDEX IF EXISTS CHARTEVENTS_idx04;
+CREATE INDEX ICHARTEVENTS_idx04
+ ON CHARTEVENTS (ICUSTAY_ID);
+
+DROP INDEX IF EXISTS CHARTEVENTS_idx05;
+CREATE INDEX CHARTEVENTS_idx05
+ ON CHARTEVENTS (ITEMID);
+
+---------------
+-- CPTEVENTS
+---------------
+
+DROP INDEX IF EXISTS CPTEVENTS_idx01;
+CREATE INDEX CPTEVENTS_idx01
+ ON CPTEVENTS (SUBJECT_ID);
+
+DROP INDEX IF EXISTS CPTEVENTS_idx02;
+CREATE INDEX CPTEVENTS_idx02
+ ON CPTEVENTS (CPT_CD);
+
+-----------
+-- D_CPT
+-----------
+
+-- Table is 134 rows - doesn't need an index.
+
+--------------------
+-- D_ICD_DIAGNOSES
+--------------------
+
+DROP INDEX IF EXISTS D_ICD_DIAG_idx01;
+CREATE INDEX D_ICD_DIAG_idx01
+ ON D_ICD_DIAGNOSES (ICD9_CODE);
+
+DROP INDEX IF EXISTS D_ICD_DIAG_idx02;
+CREATE INDEX D_ICD_DIAG_idx02
+ ON D_ICD_DIAGNOSES (LONG_TITLE);
+
+--------------------
+-- D_ICD_PROCEDURES
+--------------------
+
+DROP INDEX IF EXISTS D_ICD_PROC_idx01;
+CREATE INDEX D_ICD_PROC_idx01
+ ON D_ICD_PROCEDURES (ICD9_CODE);
+
+DROP INDEX IF EXISTS D_ICD_PROC_idx02;
+CREATE INDEX D_ICD_PROC_idx02
+ ON D_ICD_PROCEDURES (LONG_TITLE);
+
+-----------
+-- D_ITEMS
+-----------
+
+DROP INDEX IF EXISTS D_ITEMS_idx01;
+CREATE INDEX D_ITEMS_idx01
+ ON D_ITEMS (ITEMID);
+
+DROP INDEX IF EXISTS D_ITEMS_idx02;
+CREATE INDEX D_ITEMS_idx02
+ ON D_ITEMS (LABEL);
+
+-- DROP INDEX IF EXISTS D_ITEMS_idx03;
+-- CREATE INDEX D_ITEMS_idx03
+-- ON D_ITEMS (CATEGORY);
+
+---------------
+-- D_LABITEMS
+---------------
+
+DROP INDEX IF EXISTS D_LABITEMS_idx01;
+CREATE INDEX D_LABITEMS_idx01
+ ON D_LABITEMS (ITEMID);
+
+DROP INDEX IF EXISTS D_LABITEMS_idx02;
+CREATE INDEX D_LABITEMS_idx02
+ ON D_LABITEMS (LABEL);
+
+DROP INDEX IF EXISTS D_LABITEMS_idx03;
+CREATE INDEX D_LABITEMS_idx03
+ ON D_LABITEMS (LOINC_CODE);
+
+-------------------
+-- DATETIMEEVENTS
+-------------------
+
+DROP INDEX IF EXISTS DATETIMEEVENTS_idx01;
+CREATE INDEX DATETIMEEVENTS_idx01
+ ON DATETIMEEVENTS (SUBJECT_ID);
+
+DROP INDEX IF EXISTS DATETIMEEVENTS_idx02;
+CREATE INDEX DATETIMEEVENTS_idx02
+ ON DATETIMEEVENTS (ITEMID);
+
+DROP INDEX IF EXISTS DATETIMEEVENTS_idx03;
+CREATE INDEX DATETIMEEVENTS_idx03
+ ON DATETIMEEVENTS (ICUSTAY_ID);
+
+DROP INDEX IF EXISTS DATETIMEEVENTS_idx04;
+CREATE INDEX DATETIMEEVENTS_idx04
+ ON DATETIMEEVENTS (HADM_ID);
+
+-- DROP INDEX IF EXISTS DATETIMEEVENTS_idx05;
+-- CREATE INDEX DATETIMEEVENTS_idx05
+-- ON DATETIMEEVENTS (VALUE);
+
+------------------
+-- DIAGNOSES_ICD
+------------------
+
+DROP INDEX IF EXISTS DIAGNOSES_ICD_idx01;
+CREATE INDEX DIAGNOSES_ICD_idx01
+ ON DIAGNOSES_ICD (SUBJECT_ID);
+
+DROP INDEX IF EXISTS DIAGNOSES_ICD_idx02;
+CREATE INDEX DIAGNOSES_ICD_idx02
+ ON DIAGNOSES_ICD (ICD9_CODE);
+
+DROP INDEX IF EXISTS DIAGNOSES_ICD_idx03;
+CREATE INDEX DIAGNOSES_ICD_idx03
+ ON DIAGNOSES_ICD (HADM_ID);
+
+--------------
+-- DRGCODES
+--------------
+
+DROP INDEX IF EXISTS DRGCODES_idx01;
+CREATE INDEX DRGCODES_idx01
+ ON DRGCODES (SUBJECT_ID);
+
+DROP INDEX IF EXISTS DRGCODES_idx02;
+CREATE INDEX DRGCODES_idx02
+ ON DRGCODES (DRG_CODE);
+
+DROP INDEX IF EXISTS DRGCODES_idx03;
+CREATE INDEX DRGCODES_idx03
+ ON DRGCODES (DESCRIPTION);
+
+-- HADM_ID
+
+------------------
+-- ICUSTAYS
+------------------
+
+DROP INDEX IF EXISTS ICUSTAYS_idx01;
+CREATE INDEX ICUSTAYS_idx01
+ ON ICUSTAYS (SUBJECT_ID);
+
+DROP INDEX IF EXISTS ICUSTAYS_idx02;
+CREATE INDEX ICUSTAYS_idx02
+ ON ICUSTAYS (ICUSTAY_ID);
+
+-- DROP INDEX IF EXISTS ICUSTAYS_idx03;
+-- CREATE INDEX ICUSTAYS_idx03
+-- ON ICUSTAYS (LOS);
+
+-- DROP INDEX IF EXISTS ICUSTAYS_idx04;
+-- CREATE INDEX ICUSTAYS_idx04
+-- ON ICUSTAYS (FIRST_CAREUNIT);
+
+-- DROP INDEX IF EXISTS ICUSTAYS_idx05;
+-- CREATE INDEX ICUSTAYS_idx05
+-- ON ICUSTAYS (LAST_CAREUNIT);
+
+DROP INDEX IF EXISTS ICUSTAYS_idx06;
+CREATE INDEX ICUSTAYS_idx06
+ ON ICUSTAYS (HADM_ID);
+
+-------------
+-- INPUTEVENTS_CV
+-------------
+
+DROP INDEX IF EXISTS INPUTEVENTS_CV_idx01;
+CREATE INDEX INPUTEVENTS_CV_idx01
+ ON INPUTEVENTS_CV (SUBJECT_ID);
+
+ DROP INDEX IF EXISTS INPUTEVENTS_CV_idx02;
+ CREATE INDEX INPUTEVENTS_CV_idx02
+ ON INPUTEVENTS_CV (HADM_ID);
+
+DROP INDEX IF EXISTS INPUTEVENTS_CV_idx03;
+CREATE INDEX INPUTEVENTS_CV_idx03
+ ON INPUTEVENTS_CV (ICUSTAY_ID);
+
+DROP INDEX IF EXISTS INPUTEVENTS_CV_idx04;
+CREATE INDEX INPUTEVENTS_CV_idx04
+ ON INPUTEVENTS_CV (CHARTTIME);
+
+DROP INDEX IF EXISTS INPUTEVENTS_CV_idx05;
+CREATE INDEX INPUTEVENTS_CV_idx05
+ ON INPUTEVENTS_CV (ITEMID);
+
+-- DROP INDEX IF EXISTS INPUTEVENTS_CV_idx06;
+-- CREATE INDEX INPUTEVENTS_CV_idx06
+-- ON INPUTEVENTS_CV (RATE);
+
+-- DROP INDEX IF EXISTS INPUTEVENTS_CV_idx07;
+-- CREATE INDEX INPUTEVENTS_CV_idx07
+-- ON INPUTEVENTS_CV (AMOUNT);
+
+-- DROP INDEX IF EXISTS INPUTEVENTS_CV_idx08;
+-- CREATE INDEX INPUTEVENTS_CV_idx08
+-- ON INPUTEVENTS_CV (CGID);
+
+-- DROP INDEX IF EXISTS INPUTEVENTS_CV_idx09;
+-- CREATE INDEX INPUTEVENTS_CV_idx09
+-- ON INPUTEVENTS_CV (LINKORDERID, ORDERID);
+
+--------------
+-- LABEVENTS
+--------------
+
+DROP INDEX IF EXISTS LABEVENTS_idx01;
+CREATE INDEX LABEVENTS_idx01
+ ON LABEVENTS (SUBJECT_ID);
+
+DROP INDEX IF EXISTS LABEVENTS_idx02;
+CREATE INDEX LABEVENTS_idx02
+ ON LABEVENTS (HADM_ID);
+
+DROP INDEX IF EXISTS LABEVENTS_idx03;
+CREATE INDEX LABEVENTS_idx03
+ ON LABEVENTS (ITEMID);
+
+-- DROP INDEX IF EXISTS LABEVENTS_idx04;
+-- CREATE INDEX LABEVENTS_idx04
+-- ON LABEVENTS (VALUE, VALUENUM);
+
+----------------------
+-- MICROBIOLOGYEVENTS
+----------------------
+
+DROP INDEX IF EXISTS MICROBIOLOGYEVENTS_idx01;
+CREATE INDEX MICROBIOLOGYEVENTS_idx01
+ ON MICROBIOLOGYEVENTS (SUBJECT_ID);
+
+DROP INDEX IF EXISTS MICROBIOLOGYEVENTS_idx02;
+CREATE INDEX MICROBIOLOGYEVENTS_idx02
+ ON MICROBIOLOGYEVENTS (HADM_ID);
+
+-- DROP INDEX IF EXISTS MICROBIOLOGYEVENTS_idx03;
+-- CREATE INDEX MICROBIOLOGYEVENTS_idx03
+-- ON MICROBIOLOGYEVENTS (SPEC_ITEMID,
+-- ORG_ITEMID, AB_ITEMID);
+
+---------------
+-- NOTEEVENTS
+---------------
+
+DROP INDEX IF EXISTS NOTEEVENTS_idx01;
+CREATE INDEX NOTEEVENTS_idx01
+ ON NOTEEVENTS (SUBJECT_ID);
+
+DROP INDEX IF EXISTS NOTEEVENTS_idx02;
+CREATE INDEX NOTEEVENTS_idx02
+ ON NOTEEVENTS (HADM_ID);
+
+-- DROP INDEX IF EXISTS NOTEEVENTS_idx03;
+-- CREATE INDEX NOTEEVENTS_idx03
+-- ON NOTEEVENTS (CGID);
+
+-- DROP INDEX IF EXISTS NOTEEVENTS_idx04;
+-- CREATE INDEX NOTEEVENTS_idx04
+-- ON NOTEEVENTS (RECORD_ID);
+
+DROP INDEX IF EXISTS NOTEEVENTS_idx05;
+CREATE INDEX NOTEEVENTS_idx05
+ ON NOTEEVENTS (CATEGORY);
+
+---------------
+-- OUTPUTEVENTS
+---------------
+DROP INDEX IF EXISTS OUTPUTEVENTS_idx01;
+CREATE INDEX OUTPUTEVENTS_idx01
+ ON OUTPUTEVENTS (SUBJECT_ID);
+
+
+DROP INDEX IF EXISTS OUTPUTEVENTS_idx02;
+CREATE INDEX OUTPUTEVENTS_idx02
+ ON OUTPUTEVENTS (ITEMID);
+
+
+DROP INDEX IF EXISTS OUTPUTEVENTS_idx03;
+CREATE INDEX OUTPUTEVENTS_idx03
+ ON OUTPUTEVENTS (ICUSTAY_ID);
+
+
+DROP INDEX IF EXISTS OUTPUTEVENTS_idx04;
+CREATE INDEX OUTPUTEVENTS_idx04
+ ON OUTPUTEVENTS (HADM_ID);
+
+-- Perhaps not useful to index on just value? Index just for popular subset?
+-- DROP INDEX IF EXISTS OUTPUTEVENTS_idx05;
+-- CREATE INDEX OUTPUTEVENTS_idx05
+-- ON OUTPUTEVENTS (VALUE);
+
+-------------
+-- PATIENTS
+-------------
+
+-- Note that SUBJECT_ID is already indexed as it is unique
+
+-- DROP INDEX IF EXISTS PATIENTS_idx01;
+-- CREATE INDEX PATIENTS_idx01
+-- ON PATIENTS (EXPIRE_FLAG);
+
+------------------
+-- PRESCRIPTIONS
+------------------
+
+DROP INDEX IF EXISTS PRESCRIPTIONS_idx01;
+CREATE INDEX PRESCRIPTIONS_idx01
+ ON PRESCRIPTIONS (SUBJECT_ID);
+
+DROP INDEX IF EXISTS PRESCRIPTIONS_idx02;
+CREATE INDEX PRESCRIPTIONS_idx02
+ ON PRESCRIPTIONS (ICUSTAY_ID);
+
+DROP INDEX IF EXISTS PRESCRIPTIONS_idx03;
+CREATE INDEX PRESCRIPTIONS_idx03
+ ON PRESCRIPTIONS (DRUG_TYPE);
+
+DROP INDEX IF EXISTS PRESCRIPTIONS_idx04;
+CREATE INDEX PRESCRIPTIONS_idx04
+ ON PRESCRIPTIONS (DRUG);
+
+DROP INDEX IF EXISTS PRESCRIPTIONS_idx05;
+CREATE INDEX PRESCRIPTIONS_idx05
+ ON PRESCRIPTIONS (HADM_ID);
+
+-------------------
+-- PROCEDURES_ICD
+-------------------
+
+DROP INDEX IF EXISTS PROCEDURES_ICD_idx01;
+CREATE INDEX PROCEDURES_ICD_idx01
+ ON PROCEDURES_ICD (SUBJECT_ID);
+
+DROP INDEX IF EXISTS PROCEDURES_ICD_idx02;
+CREATE INDEX PROCEDURES_ICD_idx02
+ ON PROCEDURES_ICD (ICD9_CODE);
+
+DROP INDEX IF EXISTS PROCEDURES_ICD_idx03;
+CREATE INDEX PROCEDURES_ICD_idx03
+ ON PROCEDURES_ICD (HADM_ID);
+
+-------------
+-- SERVICES
+-------------
+
+DROP INDEX IF EXISTS SERVICES_idx01;
+CREATE INDEX SERVICES_idx01
+ ON SERVICES (SUBJECT_ID);
+
+DROP INDEX IF EXISTS SERVICES_idx02;
+CREATE INDEX SERVICES_idx02
+ ON SERVICES (HADM_ID);
+
+-- DROP INDEX IF EXISTS SERVICES_idx03;
+-- CREATE INDEX SERVICES_idx03
+-- ON SERVICES (CURR_SERVICE, PREV_SERVICE);
+
+-------------
+-- TRANSFERS
+-------------
+
+DROP INDEX IF EXISTS TRANSFERS_idx01;
+CREATE INDEX TRANSFERS_idx01
+ ON TRANSFERS (SUBJECT_ID);
+
+DROP INDEX IF EXISTS TRANSFERS_idx02;
+CREATE INDEX TRANSFERS_idx02
+ ON TRANSFERS (ICUSTAY_ID);
+
+DROP INDEX IF EXISTS TRANSFERS_idx03;
+CREATE INDEX TRANSFERS_idx03
+ ON TRANSFERS (HADM_ID);
+
+-- DROP INDEX IF EXISTS TRANSFERS_idx04;
+-- CREATE INDEX TRANSFERS_idx04
+-- ON TRANSFERS (INTIME, OUTTIME);
+
+-- DROP INDEX IF EXISTS TRANSFERS_idx05;
+-- CREATE INDEX TRANSFERS_idx05
+-- ON TRANSFERS (LOS);
diff --git a/mimic-iii-carevue/code/buildmimic/04_add_constraints.sql b/mimic-iii-carevue/code/buildmimic/04_add_constraints.sql
new file mode 100644
index 000000000..f7da734f8
--- /dev/null
+++ b/mimic-iii-carevue/code/buildmimic/04_add_constraints.sql
@@ -0,0 +1,449 @@
+-- ----------------------------------------------------------------
+--
+-- This is a script to add the MIMIC-III constraints for Postgres.
+--
+-- ----------------------------------------------------------------
+
+-- If running scripts individually, you can set the schema where all tables are created as follows:
+-- SET search_path TO mimiciii;
+
+--------------
+--ADMISSIONS--
+--------------
+
+-- subject_id
+ALTER TABLE ADMISSIONS DROP CONSTRAINT IF EXISTS admissions_fk_subject_id;
+ALTER TABLE ADMISSIONS
+ADD CONSTRAINT admissions_fk_subject_id
+ FOREIGN KEY (SUBJECT_ID)
+ REFERENCES PATIENTS(SUBJECT_ID);
+
+-----------
+--CALLOUT--
+-----------
+
+-- subject_id
+ALTER TABLE CALLOUT DROP CONSTRAINT IF EXISTS callout_fk_subject_id;
+ALTER TABLE CALLOUT
+ADD CONSTRAINT callout_fk_subject_id
+ FOREIGN KEY (SUBJECT_ID)
+ REFERENCES PATIENTS(SUBJECT_ID);
+
+-- hadm_id
+ALTER TABLE CALLOUT DROP CONSTRAINT IF EXISTS callout_fk_hadm_id;
+ALTER TABLE CALLOUT
+ADD CONSTRAINT callout_fk_hadm_id
+ FOREIGN KEY (HADM_ID)
+ REFERENCES ADMISSIONS(HADM_ID);
+
+---------------
+--CHARTEVENTS--
+---------------
+
+-- subject_id
+ALTER TABLE CHARTEVENTS DROP CONSTRAINT IF EXISTS chartevents_fk_subject_id;
+ALTER TABLE CHARTEVENTS
+ADD CONSTRAINT chartevents_fk_subject_id
+ FOREIGN KEY (SUBJECT_ID)
+ REFERENCES PATIENTS(SUBJECT_ID);
+
+-- cgid
+ALTER TABLE CHARTEVENTS DROP CONSTRAINT IF EXISTS chartevents_fk_cgid;
+ALTER TABLE CHARTEVENTS
+ADD CONSTRAINT chartevents_fk_cgid
+ FOREIGN KEY (CGID)
+ REFERENCES CAREGIVERS(CGID);
+
+-- hadm_id
+ALTER TABLE CHARTEVENTS DROP CONSTRAINT IF EXISTS chartevents_fk_hadm_id;
+ALTER TABLE CHARTEVENTS
+ADD CONSTRAINT chartevents_fk_hadm_id
+ FOREIGN KEY (HADM_ID)
+ REFERENCES ADMISSIONS(HADM_ID);
+
+-- item_id
+ALTER TABLE CHARTEVENTS DROP CONSTRAINT IF EXISTS chartevents_fk_itemid;
+ALTER TABLE CHARTEVENTS
+ADD CONSTRAINT chartevents_fk_itemid
+ FOREIGN KEY (ITEMID)
+ REFERENCES D_ITEMS(ITEMID);
+
+-- icustay_id
+ALTER TABLE CHARTEVENTS DROP CONSTRAINT IF EXISTS chartevents_fk_icustay_id;
+ALTER TABLE CHARTEVENTS
+ADD CONSTRAINT chartevents_fk_icustay_id
+ FOREIGN KEY (ICUSTAY_ID)
+ REFERENCES ICUSTAYS(ICUSTAY_ID);
+
+-------------
+--CPTEVENTS--
+-------------
+
+-- subject_id
+ALTER TABLE CPTEVENTS DROP CONSTRAINT IF EXISTS cptevents_fk_subject_id;
+ALTER TABLE CPTEVENTS
+ADD CONSTRAINT cptevents_fk_subject_id
+ FOREIGN KEY (SUBJECT_ID)
+ REFERENCES PATIENTS(SUBJECT_ID);
+
+-- hadm_id
+ALTER TABLE CPTEVENTS DROP CONSTRAINT IF EXISTS cptevents_fk_hadm_id;
+ALTER TABLE CPTEVENTS
+ADD CONSTRAINT cptevents_fk_hadm_id
+ FOREIGN KEY (HADM_ID)
+ REFERENCES ADMISSIONS(HADM_ID);
+
+------------------
+--DATETIMEEVENTS--
+------------------
+
+-- subject_id
+ALTER TABLE DATETIMEEVENTS DROP CONSTRAINT IF EXISTS datetimeevents_fk_subject_id;
+ALTER TABLE DATETIMEEVENTS
+ADD CONSTRAINT datetimeevents_fk_subject_id
+ FOREIGN KEY (SUBJECT_ID)
+ REFERENCES PATIENTS(SUBJECT_ID);
+
+-- cgid
+ALTER TABLE DATETIMEEVENTS DROP CONSTRAINT IF EXISTS datetimeevents_fk_cgid;
+ALTER TABLE DATETIMEEVENTS
+ADD CONSTRAINT datetimeevents_fk_cgid
+ FOREIGN KEY (CGID)
+ REFERENCES CAREGIVERS(CGID);
+
+-- hadm_id
+ALTER TABLE DATETIMEEVENTS DROP CONSTRAINT IF EXISTS datetimeevents_fk_hadm_id;
+ALTER TABLE DATETIMEEVENTS
+ADD CONSTRAINT datetimeevents_fk_hadm_id
+ FOREIGN KEY (HADM_ID)
+ REFERENCES ADMISSIONS(HADM_ID);
+
+-- item_id
+ALTER TABLE DATETIMEEVENTS DROP CONSTRAINT IF EXISTS datetimeevents_fk_itemid;
+ALTER TABLE DATETIMEEVENTS
+ADD CONSTRAINT datetimeevents_fk_itemid
+ FOREIGN KEY (ITEMID)
+ REFERENCES D_ITEMS(ITEMID);
+
+-- icustay_id
+ALTER TABLE DATETIMEEVENTS DROP CONSTRAINT IF EXISTS datetimeevents_fk_icustay_id;
+ALTER TABLE DATETIMEEVENTS
+ADD CONSTRAINT datetimeevents_fk_icustay_id
+ FOREIGN KEY (ICUSTAY_ID)
+ REFERENCES ICUSTAYS(ICUSTAY_ID);
+
+
+-----------------
+--DIAGNOSES_ICD--
+-----------------
+
+-- subject_id
+ALTER TABLE DIAGNOSES_ICD DROP CONSTRAINT IF EXISTS diagnoses_icd_fk_subject_id;
+ALTER TABLE DIAGNOSES_ICD
+ADD CONSTRAINT diagnoses_icd_fk_subject_id
+ FOREIGN KEY (SUBJECT_ID)
+ REFERENCES PATIENTS(SUBJECT_ID);
+
+-- hadm_id
+ALTER TABLE DIAGNOSES_ICD DROP CONSTRAINT IF EXISTS diagnoses_icd_fk_hadm_id;
+ALTER TABLE DIAGNOSES_ICD
+ADD CONSTRAINT diagnoses_icd_fk_hadm_id
+ FOREIGN KEY (HADM_ID)
+ REFERENCES ADMISSIONS(HADM_ID);
+
+-- -- ICD9_code
+-- ALTER TABLE DIAGNOSES_ICD DROP CONSTRAINT IF EXISTS diagnoses_icd_fk_icd9;
+-- ALTER TABLE DIAGNOSES_ICD
+-- ADD CONSTRAINT diagnoses_icd_fk_icd9
+-- FOREIGN KEY (ICD9_CODE)
+-- REFERENCES D_ICD_DIAGNOSES(ICD9_CODE);
+
+--------------
+---DRGCODES---
+--------------
+
+-- subject_id
+ALTER TABLE DRGCODES DROP CONSTRAINT IF EXISTS drgcodes_fk_subject_id;
+ALTER TABLE DRGCODES
+ADD CONSTRAINT drgcodes_fk_subject_id
+ FOREIGN KEY (SUBJECT_ID)
+ REFERENCES PATIENTS(SUBJECT_ID);
+
+-- hadm_id
+ALTER TABLE DRGCODES DROP CONSTRAINT IF EXISTS drgcodes_fk_hadm_id;
+ALTER TABLE DRGCODES
+ADD CONSTRAINT drgcodes_fk_hadm_id
+ FOREIGN KEY (HADM_ID)
+ REFERENCES ADMISSIONS(HADM_ID);
+
+-----------------
+--ICUSTAYS--
+-----------------
+
+-- subject_id
+ALTER TABLE ICUSTAYS DROP CONSTRAINT IF EXISTS icustays_fk_subject_id;
+ALTER TABLE ICUSTAYS
+ADD CONSTRAINT icustays_fk_subject_id
+ FOREIGN KEY (SUBJECT_ID)
+ REFERENCES PATIENTS(SUBJECT_ID);
+
+-- hadm_id
+ALTER TABLE ICUSTAYS DROP CONSTRAINT IF EXISTS icustays_fk_hadm_id;
+ALTER TABLE ICUSTAYS
+ADD CONSTRAINT icustays_fk_hadm_id
+ FOREIGN KEY (HADM_ID)
+ REFERENCES ADMISSIONS(HADM_ID);
+
+
+------------------
+--INPUTEVENTS_CV--
+------------------
+
+-- subject_id
+ALTER TABLE INPUTEVENTS_CV DROP CONSTRAINT IF EXISTS inputevents_cv_fk_subject_id;
+ALTER TABLE INPUTEVENTS_CV
+ADD CONSTRAINT inputevents_cv_fk_subject_id
+ FOREIGN KEY (SUBJECT_ID)
+ REFERENCES PATIENTS(SUBJECT_ID);
+
+-- hadm_id
+ALTER TABLE INPUTEVENTS_CV DROP CONSTRAINT IF EXISTS inputevents_cv_fk_hadm_id;
+ALTER TABLE INPUTEVENTS_CV
+ADD CONSTRAINT inputevents_cv_fk_hadm_id
+ FOREIGN KEY (HADM_ID)
+ REFERENCES ADMISSIONS(HADM_ID);
+
+-- icustay_id
+ALTER TABLE INPUTEVENTS_CV DROP CONSTRAINT IF EXISTS inputevents_cv_fk_icustay_id;
+ALTER TABLE INPUTEVENTS_CV
+ADD CONSTRAINT inputevents_cv_fk_icustay_id
+ FOREIGN KEY (ICUSTAY_ID)
+ REFERENCES ICUSTAYS(ICUSTAY_ID);
+
+-- cgid
+ALTER TABLE INPUTEVENTS_CV DROP CONSTRAINT IF EXISTS inputevents_cv_fk_cgid;
+ALTER TABLE INPUTEVENTS_CV
+ADD CONSTRAINT inputevents_cv_fk_cgid
+ FOREIGN KEY (CGID)
+ REFERENCES CAREGIVERS(CGID);
+
+-------------
+--LABEVENTS--
+-------------
+
+-- subject_id
+ALTER TABLE LABEVENTS DROP CONSTRAINT IF EXISTS labevents_fk_subject_id;
+ALTER TABLE LABEVENTS
+ADD CONSTRAINT labevents_fk_subject_id
+ FOREIGN KEY (SUBJECT_ID)
+ REFERENCES PATIENTS(SUBJECT_ID);
+
+-- hadm_id
+ALTER TABLE LABEVENTS DROP CONSTRAINT IF EXISTS labevents_fk_hadm_id;
+ALTER TABLE LABEVENTS
+ADD CONSTRAINT labevents_fk_hadm_id
+ FOREIGN KEY (HADM_ID)
+ REFERENCES ADMISSIONS(HADM_ID);
+
+-- item_id
+ALTER TABLE LABEVENTS DROP CONSTRAINT IF EXISTS labevents_fk_itemid;
+ALTER TABLE LABEVENTS
+ADD CONSTRAINT labevents_fk_itemid
+ FOREIGN KEY (ITEMID)
+ REFERENCES D_LABITEMS(ITEMID);
+
+----------------------
+--MICROBIOLOGYEVENTS--
+----------------------
+
+-- subject_id
+ALTER TABLE MICROBIOLOGYEVENTS DROP CONSTRAINT IF EXISTS microbiologyevents_fk_subject_id;
+ALTER TABLE MICROBIOLOGYEVENTS
+ADD CONSTRAINT microbiologyevents_fk_subject_id
+ FOREIGN KEY (SUBJECT_ID)
+ REFERENCES PATIENTS(SUBJECT_ID);
+
+-- hadm_id
+ALTER TABLE MICROBIOLOGYEVENTS DROP CONSTRAINT IF EXISTS microbiologyevents_fk_hadm_id;
+ALTER TABLE MICROBIOLOGYEVENTS
+ADD CONSTRAINT microbiologyevents_fk_hadm_id
+ FOREIGN KEY (HADM_ID)
+ REFERENCES ADMISSIONS(HADM_ID);
+
+-- ITEMIDs
+ALTER TABLE MICROBIOLOGYEVENTS DROP CONSTRAINT IF EXISTS microbiologyevents_fk_spec_itemid;
+ALTER TABLE MICROBIOLOGYEVENTS
+ADD CONSTRAINT microbiologyevents_fk_spec_itemid
+ FOREIGN KEY (spec_itemid)
+ REFERENCES D_ITEMS(itemid);
+ALTER TABLE MICROBIOLOGYEVENTS DROP CONSTRAINT IF EXISTS microbiologyevents_fk_org_itemid;
+ALTER TABLE MICROBIOLOGYEVENTS
+ADD CONSTRAINT microbiologyevents_fk_org_itemid
+ FOREIGN KEY (org_itemid)
+ REFERENCES D_ITEMS(itemid);
+ALTER TABLE MICROBIOLOGYEVENTS DROP CONSTRAINT IF EXISTS microbiologyevents_fk_ab_itemid;
+ALTER TABLE MICROBIOLOGYEVENTS
+ADD CONSTRAINT microbiologyevents_fk_ab_itemid
+ FOREIGN KEY (ab_itemid)
+ REFERENCES D_ITEMS(itemid);
+
+--------------
+--NOTEEVENTS--
+--------------
+
+-- subject_id
+ALTER TABLE NOTEEVENTS DROP CONSTRAINT IF EXISTS noteevents_fk_subject_id;
+ALTER TABLE NOTEEVENTS
+ADD CONSTRAINT noteevents_fk_subject_id
+ FOREIGN KEY (SUBJECT_ID)
+ REFERENCES PATIENTS(SUBJECT_ID);
+
+-- hadm_id
+ALTER TABLE NOTEEVENTS DROP CONSTRAINT IF EXISTS noteevents_fk_hadm_id;
+ALTER TABLE NOTEEVENTS
+ADD CONSTRAINT noteevents_fk_hadm_id
+ FOREIGN KEY (HADM_ID)
+ REFERENCES ADMISSIONS(HADM_ID);
+
+-- cgid
+ALTER TABLE NOTEEVENTS DROP CONSTRAINT IF EXISTS noteevents_fk_cgid;
+ALTER TABLE NOTEEVENTS
+ADD CONSTRAINT noteevents_fk_cgid
+ FOREIGN KEY (CGID)
+ REFERENCES CAREGIVERS(CGID);
+
+
+----------------
+--OUTPUTEVENTS--
+----------------
+
+-- subject_id
+ALTER TABLE OUTPUTEVENTS DROP CONSTRAINT IF EXISTS outputevents_subject_id;
+ALTER TABLE OUTPUTEVENTS
+ADD CONSTRAINT outputevents_fk_subject_id
+ FOREIGN KEY (SUBJECT_ID)
+ REFERENCES PATIENTS(SUBJECT_ID);
+
+-- hadm_id
+ALTER TABLE OUTPUTEVENTS DROP CONSTRAINT IF EXISTS outputevents_hadm_id;
+ALTER TABLE OUTPUTEVENTS
+ADD CONSTRAINT outputevents_fk_hadm_id
+ FOREIGN KEY (HADM_ID)
+ REFERENCES ADMISSIONS(HADM_ID);
+
+-- icustay_id
+ALTER TABLE OUTPUTEVENTS DROP CONSTRAINT IF EXISTS outputevents_icustay_id;
+ALTER TABLE OUTPUTEVENTS
+ADD CONSTRAINT outputevents_fk_icustay_id
+ FOREIGN KEY (ICUSTAY_ID)
+ REFERENCES ICUSTAYS(ICUSTAY_ID);
+
+-- itemid
+ALTER TABLE OUTPUTEVENTS DROP CONSTRAINT IF EXISTS outputevents_fk_itemid;
+ALTER TABLE OUTPUTEVENTS
+ADD CONSTRAINT outputevents_fk_itemid
+ FOREIGN KEY (ITEMID)
+ REFERENCES D_ITEMS(ITEMID);
+
+-- cgid
+ALTER TABLE OUTPUTEVENTS DROP CONSTRAINT IF EXISTS outputevents_cgid;
+ALTER TABLE OUTPUTEVENTS
+ADD CONSTRAINT outputevents_fk_cgid
+ FOREIGN KEY (CGID)
+ REFERENCES CAREGIVERS(CGID);
+
+
+
+-----------------
+--PRESCRIPTIONS--
+-----------------
+
+-- subject_id
+ALTER TABLE PRESCRIPTIONS DROP CONSTRAINT IF EXISTS prescriptions_fk_subject_id;
+ALTER TABLE PRESCRIPTIONS
+ADD CONSTRAINT prescriptions_fk_subject_id
+ FOREIGN KEY (SUBJECT_ID)
+ REFERENCES PATIENTS(SUBJECT_ID);
+
+-- hadm_id
+ALTER TABLE PRESCRIPTIONS DROP CONSTRAINT IF EXISTS prescriptions_fk_hadm_id;
+ALTER TABLE PRESCRIPTIONS
+ADD CONSTRAINT prescriptions_fk_hadm_id
+ FOREIGN KEY (HADM_ID)
+ REFERENCES ADMISSIONS(HADM_ID);
+
+-- icustay_id
+ALTER TABLE PRESCRIPTIONS DROP CONSTRAINT IF EXISTS prescriptions_fk_icustay_id;
+ALTER TABLE PRESCRIPTIONS
+ADD CONSTRAINT prescriptions_fk_icustay_id
+ FOREIGN KEY (ICUSTAY_ID)
+ REFERENCES ICUSTAYS(ICUSTAY_ID);
+
+------------------
+--PROCEDURES_ICD--
+------------------
+
+-- subject_id
+ALTER TABLE PROCEDURES_ICD DROP CONSTRAINT IF EXISTS procedures_icd_fk_subject_id;
+ALTER TABLE PROCEDURES_ICD
+ADD CONSTRAINT procedures_icd_fk_subject_id
+ FOREIGN KEY (SUBJECT_ID)
+ REFERENCES PATIENTS(SUBJECT_ID);
+
+-- hadm_id
+ALTER TABLE PROCEDURES_ICD DROP CONSTRAINT IF EXISTS procedures_icd_fk_hadm_id;
+ALTER TABLE PROCEDURES_ICD
+ADD CONSTRAINT procedures_icd_fk_hadm_id
+ FOREIGN KEY (HADM_ID)
+ REFERENCES ADMISSIONS(HADM_ID);
+
+-- -- ICD9_code
+-- ALTER TABLE PROCEDURES_ICD DROP CONSTRAINT IF EXISTS procedures_icd_fk_icd9;
+-- ALTER TABLE PROCEDURES_ICD
+-- ADD CONSTRAINT procedures_icd_fk_icd9
+-- FOREIGN KEY (ICD9_CODE)
+-- REFERENCES D_ICD_PROCEDURES(ICD9_CODE);
+
+------------
+--SERVICES--
+------------
+
+-- subject_id
+ALTER TABLE SERVICES DROP CONSTRAINT IF EXISTS services_fk_subject_id;
+ALTER TABLE SERVICES
+ADD CONSTRAINT services_fk_subject_id
+ FOREIGN KEY (SUBJECT_ID)
+ REFERENCES PATIENTS(SUBJECT_ID);
+
+-- hadm_id
+ALTER TABLE SERVICES DROP CONSTRAINT IF EXISTS services_fk_hadm_id;
+ALTER TABLE SERVICES
+ADD CONSTRAINT services_fk_hadm_id
+ FOREIGN KEY (HADM_ID)
+ REFERENCES ADMISSIONS(HADM_ID);
+
+-------------
+--TRANSFERS--
+-------------
+
+-- subject_id
+ALTER TABLE TRANSFERS DROP CONSTRAINT IF EXISTS transfers_fk_subject_id;
+ALTER TABLE TRANSFERS
+ADD CONSTRAINT transfers_fk_subject_id
+ FOREIGN KEY (SUBJECT_ID)
+ REFERENCES PATIENTS(SUBJECT_ID);
+
+-- hadm_id
+ALTER TABLE TRANSFERS DROP CONSTRAINT IF EXISTS transfers_fk_hadm_id;
+ALTER TABLE TRANSFERS
+ADD CONSTRAINT transfers_fk_hadm_id
+ FOREIGN KEY (HADM_ID)
+ REFERENCES ADMISSIONS(HADM_ID);
+
+-- icustay_id
+ALTER TABLE TRANSFERS DROP CONSTRAINT IF EXISTS transfers_fk_icustay_id;
+ALTER TABLE TRANSFERS
+ADD CONSTRAINT transfers_fk_icustay_id
+ FOREIGN KEY (ICUSTAY_ID)
+ REFERENCES ICUSTAYS(ICUSTAY_ID);
+
diff --git a/mimic-iii-carevue/code/buildmimic/05_checks.sql b/mimic-iii-carevue/code/buildmimic/05_checks.sql
new file mode 100644
index 000000000..1b897c967
--- /dev/null
+++ b/mimic-iii-carevue/code/buildmimic/05_checks.sql
@@ -0,0 +1,183 @@
+-- this query runs a few simple checks to make sure the database has loaded in OK
+-- These checks are designed for MIMIC-III v1.4
+
+-- If running scripts individually, you can set the schema where all tables are created as follows:
+-- SET search_path TO mimiciii;
+
+with expected as
+(
+ -- buildmimic
+ select 'admissions' as tbl, 26836 as row_count UNION ALL
+ select 'callout' as tbl, 6480 as row_count UNION ALL
+ select 'caregivers' as tbl, 7567 as row_count UNION ALL
+ select 'chartevents' as tbl, 172484058 as row_count UNION ALL
+ select 'cptevents' as tbl, 172213 as row_count UNION ALL
+ select 'd_cpt' as tbl, 134 as row_count UNION ALL
+ select 'd_icd_diagnoses' as tbl, 14567 as row_count UNION ALL
+ select 'd_icd_procedures' as tbl, 3882 as row_count UNION ALL
+ select 'd_items' as tbl, 12487 as row_count UNION ALL
+ select 'd_labitems' as tbl, 753 as row_count UNION ALL
+ select 'datetimeevents' as tbl, 1222856 as row_count UNION ALL
+ select 'diagnoses_icd' as tbl, 225345 as row_count UNION ALL
+ select 'drgcodes' as tbl, 38919 as row_count UNION ALL
+ select 'icustays' as tbl, 28391 as row_count UNION ALL
+ select 'inputevents_cv' as tbl, 13560330 as row_count UNION ALL
+ select 'labevents' as tbl, 10896310 as row_count UNION ALL
+ select 'microbiologyevents' as tbl, 281248 as row_count UNION ALL
+ select 'noteevents' as tbl, 880107 as row_count UNION ALL
+ select 'outputevents' as tbl, 2021183 as row_count UNION ALL
+ select 'patients' as tbl, 23692 as row_count UNION ALL
+ select 'prescriptions' as tbl, 1435643 as row_count UNION ALL
+ select 'procedures_icd' as tbl, 113012 as row_count UNION ALL
+ select 'services' as tbl, 32207 as row_count UNION ALL
+ select 'transfers' as tbl, 118217 as row_count UNION ALL
+ -- concepts
+ select 'code_status' as tbl, 28391 as row_count UNION ALL
+ select 'ventilation_classification' as tbl, 1388864 as row_count UNION ALL
+ select 'ventilation_durations' as tbl, 18454 as row_count UNION ALL
+ select 'crrt_durations' as tbl, 2796 as row_count UNION ALL
+ select 'dobutamine_durations' as tbl, 904 as row_count UNION ALL
+ select 'dopamine_durations' as tbl, 3420 as row_count UNION ALL
+ select 'epinephrine_durations' as tbl, 907 as row_count UNION ALL
+ select 'isuprel_durations' as tbl, 2 as row_count UNION ALL
+ select 'milrinone_durations' as tbl, 845 as row_count UNION ALL
+ select 'norepinephrine_durations' as tbl, 7092 as row_count UNION ALL
+ select 'phenylephrine_durations' as tbl, 11512 as row_count UNION ALL
+ select 'vasopressin_durations' as tbl, 1597 as row_count UNION ALL
+ select 'vasopressor_durations' as tbl, 18304 as row_count UNION ALL
+ select 'weight_durations' as tbl, 924615 as row_count UNION ALL
+ select 'elixhauser_ahrq_v37' as tbl, 26836 as row_count UNION ALL
+ select 'elixhauser_ahrq_v37_no_drg' as tbl, 26836 as row_count UNION ALL
+ select 'elixhauser_quan' as tbl, 26836 as row_count UNION ALL
+ select 'elixhauser_score_ahrq' as tbl, 26836 as row_count UNION ALL
+ select 'elixhauser_score_quan' as tbl, 26836 as row_count UNION ALL
+ select 'icustay_detail' as tbl, 28121 as row_count UNION ALL
+ select 'blood_gas_first_day' as tbl, 112259 as row_count UNION ALL
+ select 'blood_gas_first_day_arterial' as tbl, 79579 as row_count UNION ALL
+ select 'gcs_first_day' as tbl, 28391 as row_count UNION ALL
+ select 'height_first_day' as tbl, 20291 as row_count UNION ALL
+ select 'labs_first_day' as tbl, 28391 as row_count UNION ALL
+ select 'rrt_first_day' as tbl, 28391 as row_count UNION ALL
+ select 'urine_output_first_day' as tbl, 22084 as row_count UNION ALL
+ select 'ventilation_first_day' as tbl, 28391 as row_count UNION ALL
+ select 'vitals_first_day' as tbl, 5728 as row_count UNION ALL
+ select 'weight_first_day' as tbl, 20291 as row_count UNION ALL
+ select 'urine_output' as tbl, 1539085 as row_count UNION ALL
+ select 'angus' as tbl, 26836 as row_count UNION ALL
+ select 'martin' as tbl, 26836 as row_count UNION ALL
+ select 'explicit' as tbl, 26836 as row_count UNION ALL
+ select 'ccs_dx' as tbl, 15072 as row_count UNION ALL
+ select 'kdigo_creatinine' as tbl, 152444 as row_count UNION ALL
+ select 'kdigo_uo' as tbl, 1539085 as row_count UNION ALL
+ select 'kdigo_stages' as tbl, 1683340 as row_count UNION ALL
+ select 'kdigo_stages_7day' as tbl, 28391 as row_count UNION ALL
+ select 'kdigo_stages_48hr' as tbl, 28391 as row_count UNION ALL
+ select 'meld' as tbl, 28391 as row_count UNION ALL
+ select 'oasis' as tbl, 28391 as row_count UNION ALL
+ select 'sofa' as tbl, 28391 as row_count UNION ALL
+ select 'saps' as tbl, 28391 as row_count UNION ALL
+ select 'sapsii' as tbl, 28391 as row_count UNION ALL
+ select 'apsiii' as tbl, 28391 as row_count UNION ALL
+ select 'lods' as tbl, 28391 as row_count UNION ALL
+ select 'sirs' as tbl, 28391 as row_count UNION ALL
+ select 'qsofa' as tbl, 28391 as row_count UNION ALL
+ select 'sepsis3' as tbl, 28391 as row_count UNION ALL
+ select 'age' as tbl, 26836 as row_count UNION ALL
+ select 'charlson' as tbl, 26836 as row_count
+)
+
+, observed as
+(
+ -- buildmimic
+ select 'admissions' as tbl, count(*) as row_count from admissions UNION ALL
+ select 'callout' as tbl, count(*) as row_count from callout UNION ALL
+ select 'caregivers' as tbl, count(*) as row_count from caregivers UNION ALL
+ select 'chartevents' as tbl, count(*) as row_count from chartevents UNION ALL
+ select 'cptevents' as tbl, count(*) as row_count from cptevents UNION ALL
+ select 'd_cpt' as tbl, count(*) as row_count from d_cpt UNION ALL
+ select 'd_icd_diagnoses' as tbl, count(*) as row_count from d_icd_diagnoses UNION ALL
+ select 'd_icd_procedures' as tbl, count(*) as row_count from d_icd_procedures UNION ALL
+ select 'd_items' as tbl, count(*) as row_count from d_items UNION ALL
+ select 'd_labitems' as tbl, count(*) as row_count from d_labitems UNION ALL
+ select 'datetimeevents' as tbl, count(*) as row_count from datetimeevents UNION ALL
+ select 'diagnoses_icd' as tbl, count(*) as row_count from diagnoses_icd UNION ALL
+ select 'drgcodes' as tbl, count(*) as row_count from drgcodes UNION ALL
+ select 'icustays' as tbl, count(*) as row_count from icustays UNION ALL
+ select 'inputevents_cv' as tbl, count(*) as row_count from inputevents_cv UNION ALL
+ select 'labevents' as tbl, count(*) as row_count from labevents UNION ALL
+ select 'microbiologyevents' as tbl, count(*) as row_count from microbiologyevents UNION ALL
+ select 'noteevents' as tbl, count(*) as row_count from noteevents UNION ALL
+ select 'outputevents' as tbl, count(*) as row_count from outputevents UNION ALL
+ select 'patients' as tbl, count(*) as row_count from patients UNION ALL
+ select 'prescriptions' as tbl, count(*) as row_count from prescriptions UNION ALL
+ select 'procedures_icd' as tbl, count(*) as row_count from procedures_icd UNION ALL
+ select 'services' as tbl, count(*) as row_count from services UNION ALL
+ select 'transfers' as tbl, count(*) as row_count from transfers UNION ALL
+ -- concepts
+ select 'code_status' as tbl, count(*) as row_count from code_status UNION ALL
+ select 'ventilation_classification' as tbl, count(*) as row_count from ventilation_classification UNION ALL
+ select 'ventilation_durations' as tbl, count(*) as row_count from ventilation_durations UNION ALL
+ select 'crrt_durations' as tbl, count(*) as row_count from crrt_durations UNION ALL
+ select 'dobutamine_durations' as tbl, count(*) as row_count from dobutamine_durations UNION ALL
+ select 'dopamine_durations' as tbl, count(*) as row_count from dopamine_durations UNION ALL
+ select 'epinephrine_durations' as tbl, count(*) as row_count from epinephrine_durations UNION ALL
+ select 'isuprel_durations' as tbl, count(*) as row_count from isuprel_durations UNION ALL
+ select 'milrinone_durations' as tbl, count(*) as row_count from milrinone_durations UNION ALL
+ select 'norepinephrine_durations' as tbl, count(*) as row_count from norepinephrine_durations UNION ALL
+ select 'phenylephrine_durations' as tbl, count(*) as row_count from phenylephrine_durations UNION ALL
+ select 'vasopressin_durations' as tbl, count(*) as row_count from vasopressin_durations UNION ALL
+ select 'vasopressor_durations' as tbl, count(*) as row_count from vasopressor_durations UNION ALL
+ select 'weight_durations' as tbl, count(*) as row_count from weight_durations UNION ALL
+ select 'elixhauser_ahrq_v37' as tbl, count(*) as row_count from elixhauser_ahrq_v37 UNION ALL
+ select 'elixhauser_ahrq_v37_no_drg' as tbl, count(*) as row_count from elixhauser_ahrq_v37_no_drg UNION ALL
+ select 'elixhauser_quan' as tbl, count(*) as row_count from elixhauser_quan UNION ALL
+ select 'elixhauser_score_ahrq' as tbl, count(*) as row_count from elixhauser_score_ahrq UNION ALL
+ select 'elixhauser_score_quan' as tbl, count(*) as row_count from elixhauser_score_quan UNION ALL
+ select 'icustay_detail' as tbl, count(*) as row_count from icustay_detail UNION ALL
+ select 'blood_gas_first_day' as tbl, count(*) as row_count from blood_gas_first_day UNION ALL
+ select 'blood_gas_first_day_arterial' as tbl, count(*) as row_count from blood_gas_first_day_arterial UNION ALL
+ select 'gcs_first_day' as tbl, count(*) as row_count from gcs_first_day UNION ALL
+ select 'height_first_day' as tbl, count(*) as row_count from height_first_day UNION ALL
+ select 'labs_first_day' as tbl, count(*) as row_count from labs_first_day UNION ALL
+ select 'rrt_first_day' as tbl, count(*) as row_count from rrt_first_day UNION ALL
+ select 'urine_output_first_day' as tbl, count(*) as row_count from urine_output_first_day UNION ALL
+ select 'ventilation_first_day' as tbl, count(*) as row_count from ventilation_first_day UNION ALL
+ select 'vitals_first_day' as tbl, count(*) as row_count from vitals_first_day UNION ALL
+ select 'weight_first_day' as tbl, count(*) as row_count from weight_first_day UNION ALL
+ select 'urine_output' as tbl, count(*) as row_count from urine_output UNION ALL
+ select 'angus' as tbl, count(*) as row_count from angus UNION ALL
+ select 'martin' as tbl, count(*) as row_count from martin UNION ALL
+ select 'explicit' as tbl, count(*) as row_count from explicit UNION ALL
+ select 'ccs_dx' as tbl, count(*) as row_count from ccs_dx UNION ALL
+ select 'kdigo_creatinine' as tbl, count(*) as row_count from kdigo_creatinine UNION ALL
+ select 'kdigo_uo' as tbl, count(*) as row_count from kdigo_uo UNION ALL
+ select 'kdigo_stages' as tbl, count(*) as row_count from kdigo_stages UNION ALL
+ select 'kdigo_stages_7day' as tbl, count(*) as row_count from kdigo_stages_7day UNION ALL
+ select 'kdigo_stages_48hr' as tbl, count(*) as row_count from kdigo_stages_48hr UNION ALL
+ select 'meld' as tbl, count(*) as row_count from meld UNION ALL
+ select 'oasis' as tbl, count(*) as row_count from oasis UNION ALL
+ select 'sofa' as tbl, count(*) as row_count from sofa UNION ALL
+ select 'saps' as tbl, count(*) as row_count from saps UNION ALL
+ select 'sapsii' as tbl, count(*) as row_count from sapsii UNION ALL
+ select 'apsiii' as tbl, count(*) as row_count from apsiii UNION ALL
+ select 'lods' as tbl, count(*) as row_count from lods UNION ALL
+ select 'sirs' as tbl, count(*) as row_count from sirs UNION ALL
+ select 'qsofa' as tbl, count(*) as row_count from qsofa UNION ALL
+ select 'sepsis3' as tbl, count(*) as row_count from sepsis3 UNION ALL
+ select 'age' as tbl, count(*) as row_count from age UNION ALL
+ select 'charlson' as tbl, count(*) as row_count from charlson
+)
+
+select
+ exp.tbl
+ , exp.row_count as expected_count
+ , obs.row_count as observed_count
+ , case
+ when exp.row_count = obs.row_count
+ then 'PASSED'
+ else 'FAILED'
+ end as ROW_COUNT_CHECK
+from expected exp
+inner join observed obs
+ on exp.tbl = obs.tbl
+order by exp.tbl;
diff --git a/mimic-iii-carevue/code/concepts/01_code_status.sql b/mimic-iii-carevue/code/concepts/01_code_status.sql
new file mode 100644
index 000000000..38cb39bd6
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/01_code_status.sql
@@ -0,0 +1,74 @@
+drop table if exists code_status; create table code_status as
+-- This query extracts:
+-- i) a patient's first code status
+-- ii) a patient's last code status
+-- iii) the time of the first entry of DNR or CMO
+
+with t1 as
+(
+ select icustay_id, charttime, value
+ -- use row number to identify first and last code status
+ , row_number() over (partition by icustay_id order by charttime) as rnfirst
+ , row_number() over (partition by icustay_id order by charttime desc) as rnlast
+
+ -- coalesce the values
+ , case
+ when value in ('Full Code','Full code') then 1
+ else 0 end as fullcode
+ , case
+ when value in ('Comfort Measures','Comfort measures only') then 1
+ else 0 end as cmo
+ , case
+ when value = 'CPR Not Indicate' then 1
+ else 0 end as dncpr -- only in CareVue, i.e. only possible for ~60-70% of patients
+ , case
+ when value in ('Do Not Intubate','DNI (do not intubate)','DNR / DNI') then 1
+ else 0 end as dni
+ , case
+ when value in ('Do Not Resuscita','DNR (do not resuscitate)','DNR / DNI') then 1
+ else 0 end as dnr
+ from chartevents
+ where itemid in (128, 223758)
+ and value is not null
+ and value != 'Other/Remarks'
+ -- exclude rows marked as error
+ and (error is null or error = 0)
+)
+select ie.subject_id, ie.hadm_id, ie.icustay_id
+ -- first recorded code status
+ , max(case when rnfirst = 1 then t1.fullcode else null end) as fullcode_first
+ , max(case when rnfirst = 1 then t1.cmo else null end) as cmo_first
+ , max(case when rnfirst = 1 then t1.dnr else null end) as dnr_first
+ , max(case when rnfirst = 1 then t1.dni else null end) as dni_first
+ , max(case when rnfirst = 1 then t1.dncpr else null end) as dncpr_first
+
+ -- last recorded code status
+ , max(case when rnlast = 1 then t1.fullcode else null end) as fullcode_last
+ , max(case when rnlast = 1 then t1.cmo else null end) as cmo_last
+ , max(case when rnlast = 1 then t1.dnr else null end) as dnr_last
+ , max(case when rnlast = 1 then t1.dni else null end) as dni_last
+ , max(case when rnlast = 1 then t1.dncpr else null end) as dncpr_last
+
+ -- were they *at any time* given a certain code status
+ , max(t1.fullcode) as fullcode
+ , max(t1.cmo) as cmo
+ , max(t1.dnr) as dnr
+ , max(t1.dni) as dni
+ , max(t1.dncpr) as dncpr
+
+ -- time until their first DNR
+ , min(case when t1.dnr = 1 then t1.charttime else null end)
+ as dnr_first_charttime
+ , min(case when t1.dni = 1 then t1.charttime else null end)
+ as dni_first_charttime
+ , min(case when t1.dncpr = 1 then t1.charttime else null end)
+ as dncpr_first_charttime
+
+ -- first code status of CMO
+ , min(case when t1.cmo = 1 then t1.charttime else null end)
+ as timecmo_chart
+
+from icustays ie
+left join t1
+ on ie.icustay_id = t1.icustay_id
+group by ie.subject_id, ie.hadm_id, ie.icustay_id, ie.intime;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/02_ventilation_classification.sql b/mimic-iii-carevue/code/concepts/02_ventilation_classification.sql
new file mode 100644
index 000000000..04de47f53
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/02_ventilation_classification.sql
@@ -0,0 +1,121 @@
+drop table if exists ventilation_classification; create table ventilation_classification as
+-- Identify The presence of a mechanical ventilation using settings
+select
+ icustay_id, charttime
+ -- case statement determining whether it is an instance of mech vent
+ , max(
+ case
+ when itemid is null or value is null then 0 -- can't have null values
+ when itemid = 720 and value != 'Other/Remarks' then 1 -- VentTypeRecorded
+ when itemid = 223848 and value != 'Other' then 1
+ when itemid = 223849 then 1 -- ventilator mode
+ when itemid = 467 and value = 'Ventilator' then 1 -- O2 delivery device == ventilator
+ when itemid in(
+ 445, 448, 449, 450, 1340, 1486, 1600, 224687 -- minute volume
+ , 639, 654, 681, 682, 683, 684,224685,224684,224686 -- tidal volume
+ , 218,436,535,444,459,224697,224695,224696,224746,224747 -- High/Low/Peak/Mean/Neg insp force ("RespPressure")
+ , 221,1,1211,1655,2000,226873,224738,224419,224750,227187 -- Insp pressure
+ , 543 -- PlateauPressure
+ , 5865,5866,224707,224709,224705,224706 -- APRV pressure
+ , 60,437,505,506,686,220339,224700 -- PEEP
+ , 3459 -- high pressure relief
+ , 501,502,503,224702 -- PCV
+ , 223,667,668,669,670,671,672 -- TCPCV
+ , 224701 -- PSVlevel
+ )
+ then 1
+ else 0
+ end) as mechvent
+ , max(
+ case
+ -- initiation of oxygen therapy indicates the ventilation has ended
+ when itemid = 226732 and value in
+ (
+ 'Nasal cannula', -- 153714 observations
+ 'Face tent', -- 24601 observations
+ 'Aerosol-cool', -- 24560 observations
+ 'Trach mask ', -- 16435 observations
+ 'High flow neb', -- 10785 observations
+ 'Non-rebreather', -- 5182 observations
+ 'Venti mask ', -- 1947 observations
+ 'Medium conc mask ', -- 1888 observations
+ 'T-piece', -- 1135 observations
+ 'High flow nasal cannula', -- 925 observations
+ 'Ultrasonic neb', -- 9 observations
+ 'Vapomist' -- 3 observations
+ ) then 1
+ when itemid = 467 and value in
+ (
+ 'Cannula', -- 278252 observations
+ 'Nasal Cannula', -- 248299 observations
+ -- 'None', -- 95498 observations
+ 'Face Tent', -- 35766 observations
+ 'Aerosol-Cool', -- 33919 observations
+ 'Trach Mask', -- 32655 observations
+ 'Hi Flow Neb', -- 14070 observations
+ 'Non-Rebreather', -- 10856 observations
+ 'Venti Mask', -- 4279 observations
+ 'Medium Conc Mask', -- 2114 observations
+ 'Vapotherm', -- 1655 observations
+ 'T-Piece', -- 779 observations
+ 'Hood', -- 670 observations
+ 'Hut', -- 150 observations
+ 'TranstrachealCat', -- 78 observations
+ 'Heated Neb', -- 37 observations
+ 'Ultrasonic Neb' -- 2 observations
+ ) then 1
+ else 0
+ end) as oxygentherapy
+ , max(
+ case
+ when itemid is null or value is null then 0
+ -- extubated indicates ventilation event has ended
+ when itemid = 640 and value = 'Extubated' then 1
+ when itemid = 640 and value = 'Self Extubation' then 1
+ else 0
+ end)
+ as extubated
+ , max(
+ case
+ when itemid is null or value is null then 0
+ when itemid = 640 and value = 'Self Extubation' then 1
+ else 0
+ end)
+ as selfextubated
+from chartevents ce
+where ce.value is not null
+ -- exclude rows marked as error
+ and (ce.error != 1 or ce.error is null)
+ and itemid in
+ (
+ -- the below are settings used to indicate ventilation
+ 720, 223849 -- vent mode
+ , 223848 -- vent type
+ , 445, 448, 449, 450, 1340, 1486, 1600, 224687 -- minute volume
+ , 639, 654, 681, 682, 683, 684,224685,224684,224686 -- tidal volume
+ , 218,436,535,444,224697,224695,224696,224746,224747 -- High/Low/Peak/Mean ("RespPressure")
+ , 221,1,1211,1655,2000,226873,224738,224419,224750,227187 -- Insp pressure
+ , 543 -- PlateauPressure
+ , 5865,5866,224707,224709,224705,224706 -- APRV pressure
+ , 60,437,505,506,686,220339,224700 -- PEEP
+ , 3459 -- high pressure relief
+ , 501,502,503,224702 -- PCV
+ , 223,667,668,669,670,671,672 -- TCPCV
+ , 224701 -- PSVlevel
+
+ -- the below are settings used to indicate extubation
+ , 640 -- extubated
+
+ -- the below indicate oxygen/NIV, i.e. the end of a mechanical vent event
+ , 468 -- O2 Delivery Device#2
+ , 469 -- O2 Delivery Mode
+ , 470 -- O2 Flow (lpm)
+ , 471 -- O2 Flow (lpm) #2
+ , 227287 -- O2 Flow (additional cannula)
+ , 226732 -- O2 Delivery Device(s)
+ , 223834 -- O2 Flow
+
+ -- used in both oxygen + vent calculation
+ , 467 -- O2 Delivery Device
+ )
+group by icustay_id, charttime;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/03_ventilation_durations.sql b/mimic-iii-carevue/code/concepts/03_ventilation_durations.sql
new file mode 100644
index 000000000..67108c1b9
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/03_ventilation_durations.sql
@@ -0,0 +1,99 @@
+drop table if exists ventilation_durations; create table ventilation_durations as
+-- This query extracts the duration of mechanical ventilation
+-- The main goal of the query is to aggregate sequential ventilator settings
+-- into single mechanical ventilation "events". The start and end time of these
+-- events can then be used for various purposes: calculating the total duration
+-- of mechanical ventilation, cross-checking values (e.g. PaO2:FiO2 on vent), etc
+
+-- The query's logic is roughly:
+-- 1) The presence of a mechanical ventilation setting starts a new ventilation event
+-- 2) Any instance of a setting in the next 8 hours continues the event
+-- 3) Certain elements end the current ventilation event
+-- a) documented extubation ends the current ventilation
+-- b) initiation of non-invasive vent and/or oxygen ends the current vent
+
+-- See the ventilation_classification.sql query for step 1 of the above.
+-- This query has the logic for converting events into durations.
+with vd0 as
+(
+ select
+ icustay_id
+ -- this carries over the previous charttime which had a mechanical ventilation event
+ , case
+ when mechvent=1 then
+ lag(charttime, 1) over (partition by icustay_id, MechVent order by charttime)
+ else null
+ end as charttime_lag
+ , charttime
+ , mechvent
+ , oxygentherapy
+ , extubated
+ , selfextubated
+ from ventilation_classification
+)
+, vd1 as
+(
+ select
+ icustay_id
+ , charttime_lag
+ , charttime
+ , mechvent
+ , oxygentherapy
+ , extubated
+ , selfextubated
+
+ -- if this is a mechanical ventilation event, we calculate the time since the last event
+ , case
+ -- if the current observation indicates mechanical ventilation is present
+ -- calculate the time since the last vent event
+ when mechvent=1 then
+ round((cast(extract(epoch from (charttime - charttime_lag))/(60*60) as numeric)), 8)
+ else null
+ end as ventduration
+
+ , lag(extubated,1) over (partition by icustay_id, case when mechvent=1 or extubated=1 then 1 else 0 end order by charttime) as extubatedlag
+
+ -- now we determine if the current mech vent event is a "new", i.e. they've just been intubated
+ , case
+ -- if there is an extubation flag, we mark any subsequent ventilation as a new ventilation event
+ --when Extubated = 1 then 0 -- extubation is *not* a new ventilation event, the *subsequent* row is
+ when
+ lag(extubated,1) over (partition by icustay_id, case when mechvent=1 or extubated=1 then 1 else 0 end order by charttime)= 1 then 1
+ -- if patient has initiated oxygen therapy, and is not currently vented, start a newvent
+ when mechvent = 0 and oxygentherapy = 1 then 1
+ -- if there is less than 8 hours between vent settings, we do not treat this as a new ventilation event
+ when charttime > charttime_lag + interval '8 hour' then 1
+ else 0
+ end as newvent
+ -- use the staging table with only vent settings from chart events
+ from vd0 ventsettings
+)
+, vd2 as
+(
+ select vd1.*
+ -- create a cumulative sum of the instances of new ventilation
+ -- this results in a monotonic integer assigned to each instance of ventilation
+ , case when mechvent=1 or extubated = 1 then
+ sum( newvent )
+ over ( partition by icustay_id order by charttime )
+ else null end as ventnum
+ --- now we convert charttime of ventilator settings into durations
+ from vd1
+)
+-- create the durations for each mechanical ventilation instance
+select
+ icustay_id
+ -- regenerate ventnum so it's sequential
+ , row_number() over (partition by icustay_id order by ventnum) as ventnum
+ , min(charttime) as starttime
+ , max(charttime) as endtime
+ , round((cast(extract(epoch from (max(charttime) - min(charttime)))/(60*60) as numeric)), 8) as duration_hours
+from vd2
+group by icustay_id, vd2.ventnum
+having min(charttime) != max(charttime)
+-- patient had to be mechanically ventilated at least once
+-- i.e. max(mechvent) should be 1
+-- this excludes a frequent situation of NIV/oxygen before intub
+-- in these cases, ventnum=0 and max(mechvent)=0, so they are ignored
+and max(mechvent) = 1
+order by icustay_id, ventnum;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/04_crrt_durations.sql b/mimic-iii-carevue/code/concepts/04_crrt_durations.sql
new file mode 100644
index 000000000..02d4d0b39
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/04_crrt_durations.sql
@@ -0,0 +1,185 @@
+drop table if exists crrt_durations; create table crrt_durations as
+with crrt_settings as
+(
+ select
+ ce.icustay_id, ce.charttime
+ , max(
+ case
+ when ce.itemid in(
+ 224149, -- Access Pressure
+ 224144, -- Blood Flow (ml/min)
+ 228004, -- Citrate (ACD-A)
+ 225183, -- Current Goal
+ 225977, -- Dialysate Fluid
+ 224154, -- Dialysate Rate
+ 224151, -- Effluent Pressure
+ 224150, -- Filter Pressure
+ 225958, -- Heparin Concentration (units/mL)
+ 224145, -- Heparin Dose (per hour)
+ 224191, -- Hourly Patient Fluid Removal
+ 228005, -- PBP (Prefilter) Replacement Rate
+ 228006, -- Post Filter Replacement Rate
+ 225976, -- Replacement Fluid
+ 224153, -- Replacement Rate
+ 224152, -- Return Pressure
+ 226457 -- Ultrafiltrate Output
+ ) then 1
+ when ce.itemid in(
+ 29, -- Access mmHg
+ 173, -- Effluent Press mmHg
+ 192, -- Filter Pressure mmHg
+ 624, -- Return Pressure mmHg
+ 79, -- Blood Flow ml/min
+ 142, -- Current Goal
+ 146, -- Dialysate Flow ml/hr
+ 611, -- Replace Rate ml/hr
+ 5683 -- Hourly PFR
+ ) then 1
+ when ce.itemid = 665 and value in ('Active','Clot Increasing','Clots Present','No Clot Present') then 1
+ when ce.itemid = 147 and value = 'Yes' then 1
+ else 0
+ end) as RRT
+ -- Below indicates that a new instance of CRRT has started
+ , max(
+ case
+ -- System Integrity
+ when ce.itemid = 224146 and value in ('New Filter','Reinitiated') then 1
+ when ce.itemid = 665 and value in ('Initiated') then 1
+ else 0
+ end ) as rrt_start
+ -- Below indicates that the current instance of CRRT has ended
+ , max(
+ case
+ -- System Integrity
+ when ce.itemid = 224146 and value in ('Discontinued','Recirculating') then 1
+ -- the only value like DC is "DC'D", use like to avoid apostrophe
+ when ce.itemid = 665 and (value = 'Clotted' or value like 'DC%') then 1
+ -- Reason for CRRT filter change
+ when ce.itemid = 225956 then 1
+ else 0
+ end ) as rrt_end
+ from chartevents ce
+ where ce.itemid in(
+ -- MetaVision ITEMIDs
+ -- Below require special handling
+ 224146, -- System Integrity
+ 225956, -- Reason for CRRT Filter Change
+ -- Below are settings which indicate CRRT is started/continuing
+ 224149, -- Access Pressure
+ 224144, -- Blood Flow (ml/min)
+ 228004, -- Citrate (ACD-A)
+ 225183, -- Current Goal
+ 225977, -- Dialysate Fluid
+ 224154, -- Dialysate Rate
+ 224151, -- Effluent Pressure
+ 224150, -- Filter Pressure
+ 225958, -- Heparin Concentration (units/mL)
+ 224145, -- Heparin Dose (per hour)
+ 224191, -- Hourly Patient Fluid Removal
+ 228005, -- PBP (Prefilter) Replacement Rate
+ 228006, -- Post Filter Replacement Rate
+ 225976, -- Replacement Fluid
+ 224153, -- Replacement Rate
+ 224152, -- Return Pressure
+ 226457, -- Ultrafiltrate Output
+ -- CareVue ITEMIDs
+ -- Below require special handling
+ 665, -- System integrity
+ 147, -- Dialysate Infusing
+ 612, -- Replace.Fluid Infuse
+ -- Below are settings which indicate CRRT is started/continuing
+ 29, -- Access mmHg
+ 173, -- Effluent Press mmHg
+ 192, -- Filter Pressure mmHg
+ 624, -- Return Pressure mmHg
+ 142, -- Current Goal
+ 79, -- Blood Flow ml/min
+ 146, -- Dialysate Flow ml/hr
+ 611, -- Replace Rate ml/hr
+ 5683 -- Hourly PFR
+ )
+ and ce.value is not null
+ and coalesce(ce.valuenum,1) != 0 -- non-zero rates/values
+ group by icustay_id, charttime
+)
+-- create various lagged variables for future query
+, vd_lag as
+(
+ select
+ icustay_id
+ -- this carries over the previous charttime
+ , lag(charttime, 1) over w as charttime_prev_row
+ , charttime
+ , rrt
+ , rrt_start
+ , rrt_end
+ , lag(rrt_end, 1) over w as rrt_ended_prev_row
+ from crrt_settings
+ window w as
+ (
+ partition by icustay_id, case when rrt=1 or rrt_end=1 then 1 else 0 end
+ order by charttime
+ )
+)
+, vd1 as
+(
+ select
+ icustay_id
+ , charttime
+ , rrt
+ , rrt_start
+ , rrt_end
+
+ -- now we determine if the current event is a new instantiation
+ , case
+ when rrt_start = 1 then 1
+ -- if there is an end flag, we mark any subsequent event as new
+ when rrt_end = 1
+ -- note the end is *not* a new event, the *subsequent* row is
+ -- so here we output 0
+ then 0
+ when rrt_ended_prev_row = 1 then 1
+ -- if there is less than 2 hours between CRRT settings, we do not treat this as a new CRRT event
+ when round((cast(extract(epoch from (charttime - charttime_prev_row))/(60*60) as numeric)), 8) <= 2 then 0
+ else 1
+ end as newcrrt
+ -- use the temp table with only settings from chartevents
+ from vd_lag
+)
+, vd2 as
+(
+ select
+ vd1.*
+ -- create a cumulative sum of the instances of new CRRT
+ -- this results in a monotonically increasing integer assigned to each CRRT
+ , case
+ when rrt_start = 1 or rrt=1 or rrt_end = 1 then sum(newcrrt) over (partition by icustay_id order by charttime)
+ else null
+ end as num
+ --- now we convert CHARTTIME of CRRT settings into durations
+ from vd1
+ -- now we can isolate to just rows with settings
+ -- (before we had rows with start/end flags)
+ -- this removes any null values for NewCRRT
+ where
+ rrt_start = 1 or rrt = 1 or rrt_end = 1
+)
+-- create the durations for each CRRT instance
+, fin as
+(
+ select icustay_id
+ , num
+ , min(charttime) as starttime
+ , max(charttime) as endtime
+ , round((cast(extract(epoch from (max(charttime) - min(charttime)))/(60*60) as numeric)), 8) as duration_hours
+ -- add durations
+ from vd2
+ group by icustay_id, num
+ having min(charttime) != max(charttime)
+)
+select
+ icustay_id
+ , row_number() over (partition by icustay_id order by starttime) as num
+ , starttime, endtime, duration_hours
+from fin
+order by icustay_id, num;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/05_dobutamine_durations.sql b/mimic-iii-carevue/code/concepts/05_dobutamine_durations.sql
new file mode 100644
index 000000000..ea501d945
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/05_dobutamine_durations.sql
@@ -0,0 +1,206 @@
+drop table if exists dobutamine_durations; create table dobutamine_durations as
+-- This query extracts durations of dobutamine administration
+-- Consecutive administrations are numbered 1, 2, ...
+-- Total time on the drug can be calculated from this table by grouping using icustay_id
+
+-- Get drug administration data from CareVue first
+with vasocv1 as
+(
+ select
+ icustay_id, charttime
+ -- case statement determining whether the itemid is an instance of vasopressor usage
+ , max(case when itemid in (30042,30306) then 1 else 0 end) as vaso -- dobutamine
+
+ -- the 'stopped' column indicates if a vasopressor has been disconnected
+ , max(case when itemid in (30042,30306) and (stopped = 'Stopped' or stopped like 'D/C%') then 1
+ else 0 end) as vaso_stopped
+
+ , max(case when itemid in (30042,30306) and rate is not null then 1 else 0 end) as vaso_null
+ , max(case when itemid in (30042,30306) then rate else null end) as vaso_rate
+ , max(case when itemid in (30042,30306) then amount else null end) as vaso_amount
+
+ from inputevents_cv
+ where itemid in (30042,30306) -- dobutamine
+ group by icustay_id, charttime
+)
+
+, vasocv2 as
+(
+ select v.*
+ , sum(vaso_null) over (partition by icustay_id order by charttime) as vaso_partition
+ from
+ vasocv1 v
+)
+
+, vasocv3 as
+(
+ select v.*
+ , first_value(vaso_rate) over (partition by icustay_id, vaso_partition order by charttime) as vaso_prevrate_ifnull
+ from
+ vasocv2 v
+)
+
+, vasocv4 as
+(
+ select
+ icustay_id
+ , charttime
+ -- , (charttime - (lag(charttime, 1) over (partition by icustay_id, vaso order by charttime))) as delta
+
+ , vaso
+ , vaso_rate
+ , vaso_amount
+ , vaso_stopped
+ , vaso_prevrate_ifnull
+
+ -- We define start time here
+ , case
+ when vaso = 0 then null
+
+ -- if this is the first instance of the vasoactive drug
+ when vaso_rate > 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso, vaso_null
+ order by charttime
+ )
+ is null
+ then 1
+
+ -- you often get a string of 0s
+ -- we decide not to set these as 1, just because it makes vasonum sequential
+ when vaso_rate = 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 0
+ then 0
+
+ -- sometimes you get a string of null, associated with 0 volumes
+ -- same reason as before, we decide not to set these as 1
+ -- vaso_prevrate_ifnull is equal to the previous value *iff* the current value is null
+ when vaso_prevrate_ifnull = 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 0
+ then 0
+
+ -- If the last recorded rate was 0, newvaso = 1
+ when lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ ) = 0
+ then 1
+
+ -- If the last recorded vaso was D/C'd, newvaso = 1
+ when
+ lag(vaso_stopped,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 1 then 1
+
+ -- ** not sure if the below is needed
+ --when (charttime - (lag(charttime, 1) over (partition by icustay_id, vaso order by charttime))) > (interval '4 hours') then 1
+ else null
+ end as vaso_start
+ from
+ vasocv3
+)
+-- propagate start/stop flags forward in time
+
+, vasocv5 as
+(
+ select v.*
+ , sum(vaso_start) over (partition by icustay_id, vaso order by charttime) as vaso_first
+ from
+ vasocv4 v
+)
+
+, vasocv6 as
+(
+ select v.*
+ -- We define end time here
+ , case
+ when vaso = 0
+ then null
+
+ -- If the recorded vaso was D/C'd, this is an end time
+ when vaso_stopped = 1
+ then vaso_first
+
+ -- If the rate is zero, this is the end time
+ when vaso_rate = 0
+ then vaso_first
+
+ -- the last row in the table is always a potential end time
+ -- this captures patients who die/are discharged while on vasopressors
+ -- in principle, this could add an extra end time for the vasopressor
+ -- however, since we later group on vaso_start, any extra end times are ignored
+ when lead(charttime,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ ) is null
+ then vaso_first
+
+ else null
+ end as vaso_stop
+ from vasocv5 v
+)
+
+-- -- if you want to look at the results of the table before grouping:
+-- select
+-- icustay_id, charttime, vaso, vaso_rate, vaso_amount
+-- , case when vaso_stopped = 1 then 'Y' else '' end as stopped
+-- , vaso_start
+-- , vaso_first
+-- , vaso_stop
+-- from vasocv6 order by charttime;
+
+, vasocv as
+(
+ -- below groups together vasopressor administrations into groups
+ select
+ icustay_id
+ -- the first non-null rate is considered the starttime
+ , min(case when vaso_rate is not null then charttime else null end) as starttime
+ -- the *first* time the first/last flags agree is the stop time for this duration
+ , min(case when vaso_first = vaso_stop then charttime else null end) as endtime
+ from vasocv6
+ where
+ vaso_first is not null -- bogus data
+ and
+ vaso_first != 0 -- sometimes *only* a rate of 0 appears, i.e. the drug is never actually delivered
+ and
+ icustay_id is not null -- there are data for "floating" admissions, we don't worry about these
+ group by icustay_id, vaso_first
+ having -- ensure start time is not the same as end time
+ min(charttime) != min(case when vaso_first = vaso_stop then charttime else null end)
+ and
+ max(vaso_rate) > 0 -- if the rate was always 0 or null, we consider it not a real drug delivery
+)
+
+select
+ icustay_id
+ -- generate a sequential integer for convenience
+ , row_number() over (partition by icustay_id order by starttime) as vasonum
+ , starttime, endtime
+ , round((cast(extract(epoch from (endtime - starttime))/(60*60) as numeric)), 8) as duration_hours
+ -- add durations
+from
+ vasocv
+order by icustay_id, vasonum;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/06_dopamine_durations.sql b/mimic-iii-carevue/code/concepts/06_dopamine_durations.sql
new file mode 100644
index 000000000..8c5625442
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/06_dopamine_durations.sql
@@ -0,0 +1,206 @@
+drop table if exists dopamine_durations; create table dopamine_durations as
+-- This query extracts durations of dopamine administration
+-- Consecutive administrations are numbered 1, 2, ...
+-- Total time on the drug can be calculated from this table by grouping using icustay_id
+
+-- Get drug administration data from CareVue first
+with vasocv1 as
+(
+ select
+ icustay_id, charttime
+ -- case statement determining whether the ITEMID is an instance of vasopressor usage
+ , max(case when itemid in (30043,30307) then 1 else 0 end) as vaso -- dopamine
+
+ -- the 'stopped' column indicates if a vasopressor has been disconnected
+ , max(case when itemid in (30043,30307) and (stopped = 'Stopped' or stopped like 'D/C%') then 1
+ else 0 end) as vaso_stopped
+
+ , max(case when itemid in (30043,30307) and rate is not null then 1 else 0 end) as vaso_null
+ , max(case when itemid in (30043,30307) then rate else null end) as vaso_rate
+ , max(case when itemid in (30043,30307) then amount else null end) as vaso_amount
+
+ from inputevents_cv
+ where itemid in (30043,30307) -- dopamine
+ group by icustay_id, charttime
+)
+
+, vasocv2 as
+(
+ select v.*
+ , sum(vaso_null) over (partition by icustay_id order by charttime) as vaso_partition
+ from
+ vasocv1 v
+)
+
+, vasocv3 as
+(
+ select v.*
+ , first_value(vaso_rate) over (partition by icustay_id, vaso_partition order by charttime) as vaso_prevrate_ifnull
+ from
+ vasocv2 v
+)
+
+, vasocv4 as
+(
+ select
+ icustay_id
+ , charttime
+ -- , (charttime - (lag(charttime, 1) over (partition by icustay_id, vaso order by charttime))) as delta
+
+ , vaso
+ , vaso_rate
+ , vaso_amount
+ , vaso_stopped
+ , vaso_prevrate_ifnull
+
+ -- We define start time here
+ , case
+ when vaso = 0 then null
+
+ -- if this is the first instance of the vasoactive drug
+ when vaso_rate > 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso, vaso_null
+ order by charttime
+ )
+ is null
+ then 1
+
+ -- you often get a string of 0s
+ -- we decide not to set these as 1, just because it makes vasonum sequential
+ when vaso_rate = 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 0
+ then 0
+
+ -- sometimes you get a string of null, associated with 0 volumes
+ -- same reason as before, we decide not to set these as 1
+ -- vaso_prevrate_ifnull is equal to the previous value *iff* the current value is null
+ when vaso_prevrate_ifnull = 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 0
+ then 0
+
+ -- If the last recorded rate was 0, newvaso = 1
+ when lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ ) = 0
+ then 1
+
+ -- If the last recorded vaso was D/C'd, newvaso = 1
+ when
+ lag(vaso_stopped,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 1 then 1
+
+ -- ** not sure if the below is needed
+ --when (charttime - (lag(charttime, 1) over (partition by icustay_id, vaso order by charttime))) > (interval '4 hours') then 1
+ else null
+ end as vaso_start
+ from
+ vasocv3
+)
+-- propagate start/stop flags forward in time
+
+, vasocv5 as
+(
+ select v.*
+ , sum(vaso_start) over (partition by icustay_id, vaso order by charttime) as vaso_first
+ from
+ vasocv4 v
+)
+
+, vasocv6 as
+(
+ select v.*
+ -- We define end time here
+ , case
+ when vaso = 0
+ then null
+
+ -- If the recorded vaso was D/C'd, this is an end time
+ when vaso_stopped = 1
+ then vaso_first
+
+ -- If the rate is zero, this is the end time
+ when vaso_rate = 0
+ then vaso_first
+
+ -- the last row in the table is always a potential end time
+ -- this captures patients who die/are discharged while on vasopressors
+ -- in principle, this could add an extra end time for the vasopressor
+ -- however, since we later group on vaso_start, any extra end times are ignored
+ when lead(charttime,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ ) is null
+ then vaso_first
+
+ else null
+ end as vaso_stop
+ from vasocv5 v
+)
+
+-- -- if you want to look at the results of the table before grouping:
+-- select
+-- icustay_id, charttime, vaso, vaso_rate, vaso_amount
+-- , case when vaso_stopped = 1 then 'Y' else '' end as stopped
+-- , vaso_start
+-- , vaso_first
+-- , vaso_stop
+-- from vasocv6 order by charttime;
+
+, vasocv as
+(
+ -- below groups together vasopressor administrations into groups
+ select
+ icustay_id
+ -- the first non-null rate is considered the starttime
+ , min(case when vaso_rate is not null then charttime else null end) as starttime
+ -- the *first* time the first/last flags agree is the stop time for this duration
+ , min(case when vaso_first = vaso_stop then charttime else null end) as endtime
+ from vasocv6
+ where
+ vaso_first is not null -- bogus data
+ and
+ vaso_first != 0 -- sometimes *only* a rate of 0 appears, i.e. the drug is never actually delivered
+ and
+ icustay_id is not null -- there are data for "floating" admissions, we don't worry about these
+ group by icustay_id, vaso_first
+ having -- ensure start time is not the same as end time
+ min(charttime) != min(case when vaso_first = vaso_stop then charttime else null end)
+ and
+ max(vaso_rate) > 0 -- if the rate was always 0 or null, we consider it not a real drug delivery
+)
+
+select
+ icustay_id
+ -- generate a sequential integer for convenience
+ , row_number() over (partition by icustay_id order by starttime) as vasonum
+ , starttime, endtime
+ , round((cast(extract(epoch from (endtime - starttime))/(60*60) as numeric)), 8) as duration_hours
+ -- add durations
+from
+ vasocv
+order by icustay_id, vasonum;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/07_epinephrine_durations.sql b/mimic-iii-carevue/code/concepts/07_epinephrine_durations.sql
new file mode 100644
index 000000000..f2f787d57
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/07_epinephrine_durations.sql
@@ -0,0 +1,206 @@
+drop table if exists epinephrine_durations; create table epinephrine_durations as
+-- This query extracts durations of epinephrine administration
+-- Consecutive administrations are numbered 1, 2, ...
+-- Total time on the drug can be calculated from this table by grouping using icustay_id
+
+-- Get drug administration data from CareVue first
+with vasocv1 as
+(
+ select
+ icustay_id, charttime
+ -- case statement determining whether the ITEMID is an instance of vasopressor usage
+ , max(case when itemid in (30044,30119,30309) then 1 else 0 end) as vaso -- epinephrine
+
+ -- the 'stopped' column indicates if a vasopressor has been disconnected
+ , max(case when itemid in (30044,30119,30309) and (stopped = 'Stopped' or stopped like 'D/C%') then 1
+ else 0 end) as vaso_stopped
+
+ , max(case when itemid in (30044,30119,30309) and rate is not null then 1 else 0 end) as vaso_null
+ , max(case when itemid in (30044,30119,30309) then rate else null end) as vaso_rate
+ , max(case when itemid in (30044,30119,30309) then amount else null end) as vaso_amount
+
+ from inputevents_cv
+ where itemid in (30044,30119,30309) -- epinephrine
+ group by icustay_id, charttime
+)
+
+, vasocv2 as
+(
+ select v.*
+ , sum(vaso_null) over (partition by icustay_id order by charttime) as vaso_partition
+ from
+ vasocv1 v
+)
+
+, vasocv3 as
+(
+ select v.*
+ , first_value(vaso_rate) over (partition by icustay_id, vaso_partition order by charttime) as vaso_prevrate_ifnull
+ from
+ vasocv2 v
+)
+
+, vasocv4 as
+(
+ select
+ icustay_id
+ , charttime
+ -- , (charttime - (lag(charttime, 1) over (partition by icustay_id, vaso order by charttime))) as delta
+
+ , vaso
+ , vaso_rate
+ , vaso_amount
+ , vaso_stopped
+ , vaso_prevrate_ifnull
+
+ -- We define start time here
+ , case
+ when vaso = 0 then null
+
+ -- if this is the first instance of the vasoactive drug
+ when vaso_rate > 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso, vaso_null
+ order by charttime
+ )
+ is null
+ then 1
+
+ -- you often get a string of 0s
+ -- we decide not to set these as 1, just because it makes vasonum sequential
+ when vaso_rate = 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 0
+ then 0
+
+ -- sometimes you get a string of null, associated with 0 volumes
+ -- same reason as before, we decide not to set these as 1
+ -- vaso_prevrate_ifnull is equal to the previous value *iff* the current value is null
+ when vaso_prevrate_ifnull = 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 0
+ then 0
+
+ -- If the last recorded rate was 0, newvaso = 1
+ when lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ ) = 0
+ then 1
+
+ -- If the last recorded vaso was D/C'd, newvaso = 1
+ when
+ lag(vaso_stopped,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 1 then 1
+
+ -- ** not sure if the below is needed
+ --when (charttime - (lag(charttime, 1) over (partition by icustay_id, vaso order by charttime))) > (interval '4 hours') then 1
+ else null
+ end as vaso_start
+ from
+ vasocv3
+)
+-- propagate start/stop flags forward in time
+
+, vasocv5 as
+(
+ select v.*
+ , sum(vaso_start) over (partition by icustay_id, vaso order by charttime) as vaso_first
+ from
+ vasocv4 v
+)
+
+, vasocv6 as
+(
+ select v.*
+ -- We define end time here
+ , case
+ when vaso = 0
+ then null
+
+ -- If the recorded vaso was D/C'd, this is an end time
+ when vaso_stopped = 1
+ then vaso_first
+
+ -- If the rate is zero, this is the end time
+ when vaso_rate = 0
+ then vaso_first
+
+ -- the last row in the table is always a potential end time
+ -- this captures patients who die/are discharged while on vasopressors
+ -- in principle, this could add an extra end time for the vasopressor
+ -- however, since we later group on vaso_start, any extra end times are ignored
+ when lead(charttime,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ ) is null
+ then vaso_first
+
+ else null
+ end as vaso_stop
+ from vasocv5 v
+)
+
+-- -- if you want to look at the results of the table before grouping:
+-- select
+-- icustay_id, charttime, vaso, vaso_rate, vaso_amount
+-- , case when vaso_stopped = 1 then 'Y' else '' end as stopped
+-- , vaso_start
+-- , vaso_first
+-- , vaso_stop
+-- from vasocv6 order by charttime;
+
+, vasocv as
+(
+ -- below groups together vasopressor administrations into groups
+ select
+ icustay_id
+ -- the first non-null rate is considered the starttime
+ , min(case when vaso_rate is not null then charttime else null end) as starttime
+ -- the *first* time the first/last flags agree is the stop time for this duration
+ , min(case when vaso_first = vaso_stop then charttime else null end) as endtime
+ from vasocv6
+ where
+ vaso_first is not null -- bogus data
+ and
+ vaso_first != 0 -- sometimes *only* a rate of 0 appears, i.e. the drug is never actually delivered
+ and
+ icustay_id is not null -- there are data for "floating" admissions, we don't worry about these
+ group by icustay_id, vaso_first
+ having -- ensure start time is not the same as end time
+ min(charttime) != min(case when vaso_first = vaso_stop then charttime else null end)
+ and
+ max(vaso_rate) > 0 -- if the rate was always 0 or null, we consider it not a real drug delivery
+)
+
+select
+ icustay_id
+ -- generate a sequential integer for convenience
+ , row_number() over (partition by icustay_id order by starttime) as vasonum
+ , starttime, endtime
+ , round((cast(extract(epoch from (endtime - starttime))/(60*60) as numeric)), 8) as duration_hours
+ -- add durations
+from
+ vasocv
+order by icustay_id, vasonum;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/08_isuprel_durations.sql b/mimic-iii-carevue/code/concepts/08_isuprel_durations.sql
new file mode 100644
index 000000000..4f1339363
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/08_isuprel_durations.sql
@@ -0,0 +1,206 @@
+drop table if exists isuprel_durations; create table isuprel_durations as
+-- This query extracts durations of isuprel administration
+-- Consecutive administrations are numbered 1, 2, ...
+-- Total time on the drug can be calculated from this table by grouping using icustay_id
+
+-- Get drug administration data from CareVue first
+with vasocv1 as
+(
+ select
+ icustay_id, charttime
+ -- case statement determining whether the itemid is an instance of vasopressor usage
+ , max(case when itemid = 30046 then 1 else 0 end) as vaso -- Isuprel
+
+ -- the 'stopped' column indicates if a vasopressor has been disconnected
+ , max(case when itemid = 30046 and (stopped = 'Stopped' or stopped like 'D/C%') then 1
+ else 0 end) as vaso_stopped
+
+ , max(case when itemid = 30046 and rate is not null then 1 else 0 end) as vaso_null
+ , max(case when itemid = 30046 then rate else null end) as vaso_rate
+ , max(case when itemid = 30046 then amount else null end) as vaso_amount
+
+ from inputevents_cv
+ where itemid = 30046 -- isuprel
+ group by icustay_id, charttime
+)
+
+, vasocv2 as
+(
+ select v.*
+ , sum(vaso_null) over (partition by icustay_id order by charttime) as vaso_partition
+ from
+ vasocv1 v
+)
+
+, vasocv3 as
+(
+ select v.*
+ , first_value(vaso_rate) over (partition by icustay_id, vaso_partition order by charttime) as vaso_prevrate_ifnull
+ from
+ vasocv2 v
+)
+
+, vasocv4 as
+(
+ select
+ icustay_id
+ , charttime
+ -- , (charttime - (lag(charttime, 1) over (partition by icustay_id, vaso order by charttime))) as delta
+
+ , vaso
+ , vaso_rate
+ , vaso_amount
+ , vaso_stopped
+ , vaso_prevrate_ifnull
+
+ -- We define start time here
+ , case
+ when vaso = 0 then null
+
+ -- if this is the first instance of the vasoactive drug
+ when vaso_rate > 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso, vaso_null
+ order by charttime
+ )
+ is null
+ then 1
+
+ -- you often get a string of 0s
+ -- we decide not to set these as 1, just because it makes vasonum sequential
+ when vaso_rate = 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 0
+ then 0
+
+ -- sometimes you get a string of null, associated with 0 volumes
+ -- same reason as before, we decide not to set these as 1
+ -- vaso_prevrate_ifnull is equal to the previous value *iff* the current value is null
+ when vaso_prevrate_ifnull = 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 0
+ then 0
+
+ -- If the last recorded rate was 0, newvaso = 1
+ when lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ ) = 0
+ then 1
+
+ -- If the last recorded vaso was D/C'd, newvaso = 1
+ when
+ lag(vaso_stopped,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 1 then 1
+
+ -- ** not sure if the below is needed
+ --when (charttime - (lag(charttime, 1) over (partition by icustay_id, vaso order by charttime))) > (interval '4 hours') then 1
+ else null
+ end as vaso_start
+ from
+ vasocv3
+)
+-- propagate start/stop flags forward in time
+
+, vasocv5 as
+(
+ select v.*
+ , sum(vaso_start) over (partition by icustay_id, vaso order by charttime) as vaso_first
+ from
+ vasocv4 v
+)
+
+, vasocv6 as
+(
+ select v.*
+ -- We define end time here
+ , case
+ when vaso = 0
+ then null
+
+ -- If the recorded vaso was D/C'd, this is an end time
+ when vaso_stopped = 1
+ then vaso_first
+
+ -- If the rate is zero, this is the end time
+ when vaso_rate = 0
+ then vaso_first
+
+ -- the last row in the table is always a potential end time
+ -- this captures patients who die/are discharged while on vasopressors
+ -- in principle, this could add an extra end time for the vasopressor
+ -- however, since we later group on vaso_start, any extra end times are ignored
+ when lead(charttime,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ ) is null
+ then vaso_first
+
+ else null
+ end as vaso_stop
+ from vasocv5 v
+)
+
+-- -- if you want to look at the results of the table before grouping:
+-- select
+-- icustay_id, charttime, vaso, vaso_rate, vaso_amount
+-- , case when vaso_stopped = 1 then 'Y' else '' end as stopped
+-- , vaso_start
+-- , vaso_first
+-- , vaso_stop
+-- from vasocv6 order by charttime;
+
+, vasocv as
+(
+ -- below groups together vasopressor administrations into groups
+ select
+ icustay_id
+ -- the first non-null rate is considered the starttime
+ , min(case when vaso_rate is not null then charttime else null end) as starttime
+ -- the *first* time the first/last flags agree is the stop time for this duration
+ , min(case when vaso_first = vaso_stop then charttime else null end) as endtime
+ from vasocv6
+ where
+ vaso_first is not null -- bogus data
+ and
+ vaso_first != 0 -- sometimes *only* a rate of 0 appears, i.e. the drug is never actually delivered
+ and
+ icustay_id is not null -- there are data for "floating" admissions, we don't worry about these
+ group by icustay_id, vaso_first
+ having -- ensure start time is not the same as end time
+ min(charttime) != min(case when vaso_first = vaso_stop then charttime else null end)
+ and
+ max(vaso_rate) > 0 -- if the rate was always 0 or null, we consider it not a real drug delivery
+)
+
+select
+ icustay_id
+ -- generate a sequential integer for convenience
+ , row_number() over (partition by icustay_id order by starttime) as vasonum
+ , starttime, endtime
+ , round((cast(extract(epoch from (endtime - starttime))/(60*60) as numeric)), 8) as duration_hours
+ -- add durations
+from
+ vasocv
+order by icustay_id, vasonum;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/09_milrinone_durations.sql b/mimic-iii-carevue/code/concepts/09_milrinone_durations.sql
new file mode 100644
index 000000000..d7d2865af
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/09_milrinone_durations.sql
@@ -0,0 +1,206 @@
+drop table if exists milrinone_durations; create table milrinone_durations as
+-- This query extracts durations of milrinone administration
+-- Consecutive administrations are numbered 1, 2, ...
+-- Total time on the drug can be calculated from this table by grouping using icustay_id
+
+-- Get drug administration data from CareVue first
+with vasocv1 as
+(
+ select
+ icustay_id, charttime
+ -- case statement determining whether the itemid is an instance of vasopressor usage
+ , max(case when itemid = 30125 then 1 else 0 end) as vaso -- milrinone
+
+ -- the 'stopped' column indicates if a vasopressor has been disconnected
+ , max(case when itemid = 30125 and (stopped = 'Stopped' or stopped like 'D/C%') then 1
+ else 0 end) as vaso_stopped
+
+ , max(case when itemid = 30125 and rate is not null then 1 else 0 end) as vaso_null
+ , max(case when itemid = 30125 then rate else null end) as vaso_rate
+ , max(case when itemid = 30125 then amount else null end) as vaso_amount
+
+ from inputevents_cv
+ where itemid = 30125 -- milrinone
+ group by icustay_id, charttime
+)
+
+, vasocv2 as
+(
+ select v.*
+ , sum(vaso_null) over (partition by icustay_id order by charttime) as vaso_partition
+ from
+ vasocv1 v
+)
+
+, vasocv3 as
+(
+ select v.*
+ , first_value(vaso_rate) over (partition by icustay_id, vaso_partition order by charttime) as vaso_prevrate_ifnull
+ from
+ vasocv2 v
+)
+
+, vasocv4 as
+(
+ select
+ icustay_id
+ , charttime
+ -- , (charttime - (lag(charttime, 1) over (partition by icustay_id, vaso order by charttime))) as delta
+
+ , vaso
+ , vaso_rate
+ , vaso_amount
+ , vaso_stopped
+ , vaso_prevrate_ifnull
+
+ -- We define start time here
+ , case
+ when vaso = 0 then null
+
+ -- if this is the first instance of the vasoactive drug
+ when vaso_rate > 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso, vaso_null
+ order by charttime
+ )
+ is null
+ then 1
+
+ -- you often get a string of 0s
+ -- we decide not to set these as 1, just because it makes vasonum sequential
+ when vaso_rate = 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 0
+ then 0
+
+ -- sometimes you get a string of null, associated with 0 volumes
+ -- same reason as before, we decide not to set these as 1
+ -- vaso_prevrate_ifnull is equal to the previous value *iff* the current value is null
+ when vaso_prevrate_ifnull = 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 0
+ then 0
+
+ -- If the last recorded rate was 0, newvaso = 1
+ when lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ ) = 0
+ then 1
+
+ -- If the last recorded vaso was D/C'd, newvaso = 1
+ when
+ lag(vaso_stopped,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 1 then 1
+
+ -- ** not sure if the below is needed
+ --when (charttime - (lag(charttime, 1) over (partition by icustay_id, vaso order by charttime))) > (interval '4 hours') then 1
+ else null
+ end as vaso_start
+ from
+ vasocv3
+)
+-- propagate start/stop flags forward in time
+
+, vasocv5 as
+(
+ select v.*
+ , sum(vaso_start) over (partition by icustay_id, vaso order by charttime) as vaso_first
+ from
+ vasocv4 v
+)
+
+, vasocv6 as
+(
+ select v.*
+ -- We define end time here
+ , case
+ when vaso = 0
+ then null
+
+ -- If the recorded vaso was D/C'd, this is an end time
+ when vaso_stopped = 1
+ then vaso_first
+
+ -- If the rate is zero, this is the end time
+ when vaso_rate = 0
+ then vaso_first
+
+ -- the last row in the table is always a potential end time
+ -- this captures patients who die/are discharged while on vasopressors
+ -- in principle, this could add an extra end time for the vasopressor
+ -- however, since we later group on vaso_start, any extra end times are ignored
+ when lead(charttime,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ ) is null
+ then vaso_first
+
+ else null
+ end as vaso_stop
+ from vasocv5 v
+)
+
+-- -- if you want to look at the results of the table before grouping:
+-- select
+-- icustay_id, charttime, vaso, vaso_rate, vaso_amount
+-- , case when vaso_stopped = 1 then 'Y' else '' end as stopped
+-- , vaso_start
+-- , vaso_first
+-- , vaso_stop
+-- from vasocv6 order by charttime;
+
+, vasocv as
+(
+ -- below groups together vasopressor administrations into groups
+ select
+ icustay_id
+ -- the first non-null rate is considered the starttime
+ , min(case when vaso_rate is not null then charttime else null end) as starttime
+ -- the *first* time the first/last flags agree is the stop time for this duration
+ , min(case when vaso_first = vaso_stop then charttime else null end) as endtime
+ from vasocv6
+ where
+ vaso_first is not null -- bogus data
+ and
+ vaso_first != 0 -- sometimes *only* a rate of 0 appears, i.e. the drug is never actually delivered
+ and
+ icustay_id is not null -- there are data for "floating" admissions, we don't worry about these
+ group by icustay_id, vaso_first
+ having -- ensure start time is not the same as end time
+ min(charttime) != min(case when vaso_first = vaso_stop then charttime else null end)
+ and
+ max(vaso_rate) > 0 -- if the rate was always 0 or null, we consider it not a real drug delivery
+)
+
+select
+ icustay_id
+ -- generate a sequential integer for convenience
+ , row_number() over (partition by icustay_id order by starttime) as vasonum
+ , starttime, endtime
+ , round((cast(extract(epoch from (endtime - starttime))/(60*60) as numeric)), 8) as duration_hours
+ -- add durations
+from
+ vasocv
+order by icustay_id, vasonum;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/10_norepinephrine_durations.sql b/mimic-iii-carevue/code/concepts/10_norepinephrine_durations.sql
new file mode 100644
index 000000000..d3c4b3ffa
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/10_norepinephrine_durations.sql
@@ -0,0 +1,206 @@
+drop table if exists norepinephrine_durations; create table norepinephrine_durations as
+-- This query extracts durations of norepinephrine administration
+-- Consecutive administrations are numbered 1, 2, ...
+-- Total time on the drug can be calculated from this table by grouping using icustay_id
+
+-- Get drug administration data from CareVue first
+with vasocv1 as
+(
+ select
+ icustay_id, charttime
+ -- case statement determining whether the itemid is an instance of vasopressor usage
+ , max(case when itemid in (30047,30120) then 1 else 0 end) as vaso -- norepinephrine
+
+ -- the 'stopped' column indicates if a vasopressor has been disconnected
+ , max(case when itemid in (30047,30120) and (stopped = 'Stopped' or stopped like 'D/C%') then 1
+ else 0 end) as vaso_stopped
+
+ , max(case when itemid in (30047,30120) and rate is not null then 1 else 0 end) as vaso_null
+ , max(case when itemid in (30047,30120) then rate else null end) as vaso_rate
+ , max(case when itemid in (30047,30120) then amount else null end) as vaso_amount
+
+ from inputevents_cv
+ where itemid in (30047,30120) -- norepinephrine
+ group by icustay_id, charttime
+)
+
+, vasocv2 as
+(
+ select v.*
+ , sum(vaso_null) over (partition by icustay_id order by charttime) as vaso_partition
+ from
+ vasocv1 v
+)
+
+, vasocv3 as
+(
+ select v.*
+ , first_value(vaso_rate) over (partition by icustay_id, vaso_partition order by charttime) as vaso_prevrate_ifnull
+ from
+ vasocv2 v
+)
+
+, vasocv4 as
+(
+ select
+ icustay_id
+ , charttime
+ -- , (charttime - (lag(charttime, 1) over (partition by icustay_id, vaso order by charttime))) as delta
+
+ , vaso
+ , vaso_rate
+ , vaso_amount
+ , vaso_stopped
+ , vaso_prevrate_ifnull
+
+ -- We define start time here
+ , case
+ when vaso = 0 then null
+
+ -- if this is the first instance of the vasoactive drug
+ when vaso_rate > 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso, vaso_null
+ order by charttime
+ )
+ is null
+ then 1
+
+ -- you often get a string of 0s
+ -- we decide not to set these as 1, just because it makes vasonum sequential
+ when vaso_rate = 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 0
+ then 0
+
+ -- sometimes you get a string of null, associated with 0 volumes
+ -- same reason as before, we decide not to set these as 1
+ -- vaso_prevrate_ifnull is equal to the previous value *iff* the current value is null
+ when vaso_prevrate_ifnull = 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 0
+ then 0
+
+ -- If the last recorded rate was 0, newvaso = 1
+ when lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ ) = 0
+ then 1
+
+ -- If the last recorded vaso was D/C'd, newvaso = 1
+ when
+ lag(vaso_stopped,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 1 then 1
+
+ -- ** not sure if the below is needed
+ --when (charttime - (lag(charttime, 1) over (partition by icustay_id, vaso order by charttime))) > (interval '4 hours') then 1
+ else null
+ end as vaso_start
+ from
+ vasocv3
+)
+-- propagate start/stop flags forward in time
+
+, vasocv5 as
+(
+ select v.*
+ , sum(vaso_start) over (partition by icustay_id, vaso order by charttime) as vaso_first
+ from
+ vasocv4 v
+)
+
+, vasocv6 as
+(
+ select v.*
+ -- We define end time here
+ , case
+ when vaso = 0
+ then null
+
+ -- If the recorded vaso was D/C'd, this is an end time
+ when vaso_stopped = 1
+ then vaso_first
+
+ -- If the rate is zero, this is the end time
+ when vaso_rate = 0
+ then vaso_first
+
+ -- the last row in the table is always a potential end time
+ -- this captures patients who die/are discharged while on vasopressors
+ -- in principle, this could add an extra end time for the vasopressor
+ -- however, since we later group on vaso_start, any extra end times are ignored
+ when lead(charttime,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ ) is null
+ then vaso_first
+
+ else null
+ end as vaso_stop
+ from vasocv5 v
+)
+
+-- -- if you want to look at the results of the carevue data before grouping:
+-- select
+-- icustay_id, charttime, vaso, vaso_rate, vaso_amount
+-- , case when vaso_stopped = 1 then 'Y' else '' end as stopped
+-- , vaso_start
+-- , vaso_first
+-- , vaso_stop
+-- from vasocv6 order by charttime;
+
+, vasocv as
+(
+ -- below groups together vasopressor administrations into groups
+ select
+ icustay_id
+ -- the first non-null rate is considered the starttime
+ , min(case when vaso_rate is not null then charttime else null end) as starttime
+ -- the *first* time the first/last flags agree is the stop time for this duration
+ , min(case when vaso_first = vaso_stop then charttime else null end) as endtime
+ from vasocv6
+ where
+ vaso_first is not null -- bogus data
+ and
+ vaso_first != 0 -- sometimes *only* a rate of 0 appears, i.e. the drug is never actually delivered
+ and
+ icustay_id is not null -- there are data for "floating" admissions, we don't worry about these
+ group by icustay_id, vaso_first
+ having -- ensure start time is not the same as end time
+ min(charttime) != min(case when vaso_first = vaso_stop then charttime else null end)
+ and
+ max(vaso_rate) > 0 -- if the rate was always 0 or null, we consider it not a real drug delivery
+)
+
+select
+ icustay_id
+ -- generate a sequential integer for convenience
+ , row_number() over (partition by icustay_id order by starttime) as vasonum
+ , starttime, endtime
+ , round((cast(extract(epoch from (endtime - starttime))/(60*60) as numeric)), 8) as duration_hours
+ -- add durations
+from
+ vasocv
+order by icustay_id, vasonum;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/11_phenylephrine_durations.sql b/mimic-iii-carevue/code/concepts/11_phenylephrine_durations.sql
new file mode 100644
index 000000000..95d32c859
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/11_phenylephrine_durations.sql
@@ -0,0 +1,206 @@
+drop table if exists phenylephrine_durations; create table phenylephrine_durations as
+-- This query extracts durations of phenylephrine administration
+-- Consecutive administrations are numbered 1, 2, ...
+-- Total time on the drug can be calculated from this table by grouping using icustay_id
+
+-- Get drug administration data from CareVue first
+with vasocv1 as
+(
+ select
+ icustay_id, charttime
+ -- case statement determining whether the itemid is an instance of vasopressor usage
+ , max(case when itemid in (30127,30128) then 1 else 0 end) as vaso -- phenylephrine
+
+ -- the 'stopped' column indicates if a vasopressor has been disconnected
+ , max(case when itemid in (30127,30128) and (stopped = 'Stopped' or stopped like 'D/C%') then 1
+ else 0 end) as vaso_stopped
+
+ , max(case when itemid in (30127,30128) and rate is not null then 1 else 0 end) as vaso_null
+ , max(case when itemid in (30127,30128) then rate else null end) as vaso_rate
+ , max(case when itemid in (30127,30128) then amount else null end) as vaso_amount
+
+ from inputevents_cv
+ where itemid in (30127,30128) -- phenylephrine
+ group by icustay_id, charttime
+)
+
+, vasocv2 as
+(
+ select v.*
+ , sum(vaso_null) over (partition by icustay_id order by charttime) as vaso_partition
+ from
+ vasocv1 v
+)
+
+, vasocv3 as
+(
+ select v.*
+ , first_value(vaso_rate) over (partition by icustay_id, vaso_partition order by charttime) as vaso_prevrate_ifnull
+ from
+ vasocv2 v
+)
+
+, vasocv4 as
+(
+ select
+ icustay_id
+ , charttime
+ -- , (charttime - (lag(charttime, 1) over (partition by icustay_id, vaso order by charttime))) as delta
+
+ , vaso
+ , vaso_rate
+ , vaso_amount
+ , vaso_stopped
+ , vaso_prevrate_ifnull
+
+ -- We define start time here
+ , case
+ when vaso = 0 then null
+
+ -- if this is the first instance of the vasoactive drug
+ when vaso_rate > 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso, vaso_null
+ order by charttime
+ )
+ is null
+ then 1
+
+ -- you often get a string of 0s
+ -- we decide not to set these as 1, just because it makes vasonum sequential
+ when vaso_rate = 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 0
+ then 0
+
+ -- sometimes you get a string of NULL, associated with 0 volumes
+ -- same reason as before, we decide not to set these as 1
+ -- vaso_prevrate_ifnull is equal to the previous value *iff* the current value is null
+ when vaso_prevrate_ifnull = 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 0
+ then 0
+
+ -- If the last recorded rate was 0, newvaso = 1
+ when lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ ) = 0
+ then 1
+
+ -- If the last recorded vaso was D/C'd, newvaso = 1
+ when
+ lag(vaso_stopped,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 1 then 1
+
+ -- ** not sure if the below is needed
+ --when (charttime - (lag(charttime, 1) over (partition by icustay_id, vaso order by charttime))) > (interval '4 hours') then 1
+ else null
+ end as vaso_start
+ from
+ vasocv3
+)
+-- propagate start/stop flags forward in time
+
+, vasocv5 as
+(
+ select v.*
+ , sum(vaso_start) over (partition by icustay_id, vaso order by charttime) as vaso_first
+ from
+ vasocv4 v
+)
+
+, vasocv6 as
+(
+ select v.*
+ -- We define end time here
+ , case
+ when vaso = 0
+ then null
+
+ -- If the recorded vaso was D/C'd, this is an end time
+ when vaso_stopped = 1
+ then vaso_first
+
+ -- If the rate is zero, this is the end time
+ when vaso_rate = 0
+ then vaso_first
+
+ -- the last row in the table is always a potential end time
+ -- this captures patients who die/are discharged while on vasopressors
+ -- in principle, this could add an extra end time for the vasopressor
+ -- however, since we later group on vaso_start, any extra end times are ignored
+ when lead(charttime,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ ) is null
+ then vaso_first
+
+ else null
+ end as vaso_stop
+ from vasocv5 v
+)
+
+-- -- if you want to look at the results of the table before grouping:
+-- select
+-- icustay_id, charttime, vaso, vaso_rate, vaso_amount
+-- , case when vaso_stopped = 1 then 'Y' else '' end as stopped
+-- , vaso_start
+-- , vaso_first
+-- , vaso_stop
+-- from vasocv6 order by charttime;
+
+, vasocv as
+(
+ -- below groups together vasopressor administrations into groups
+ select
+ icustay_id
+ -- the first non-null rate is considered the starttime
+ , min(case when vaso_rate is not null then charttime else null end) as starttime
+ -- the *first* time the first/last flags agree is the stop time for this duration
+ , min(case when vaso_first = vaso_stop then charttime else null end) as endtime
+ from vasocv6
+ where
+ vaso_first is not null -- bogus data
+ and
+ vaso_first != 0 -- sometimes *only* a rate of 0 appears, i.e. the drug is never actually delivered
+ and
+ icustay_id is not null -- there are data for "floating" admissions, we don't worry about these
+ group by icustay_id, vaso_first
+ having -- ensure start time is not the same as end time
+ min(charttime) != min(case when vaso_first = vaso_stop then charttime else null end)
+ and
+ max(vaso_rate) > 0 -- if the rate was always 0 or null, we consider it not a real drug delivery
+)
+
+select
+ icustay_id
+ -- generate a sequential integer for convenience
+ , row_number() over (partition by icustay_id order by starttime) as vasonum
+ , starttime, endtime
+ , round((cast(extract(epoch from (endtime - starttime))/(60*60) as numeric)), 8) as duration_hours
+ -- add durations
+from
+ vasocv
+order by icustay_id, vasonum;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/12_vasopressin_durations.sql b/mimic-iii-carevue/code/concepts/12_vasopressin_durations.sql
new file mode 100644
index 000000000..3b81ee27e
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/12_vasopressin_durations.sql
@@ -0,0 +1,206 @@
+drop table if exists vasopressin_durations; create table vasopressin_durations as
+-- This query extracts durations of vasopressin administration
+-- Consecutive administrations are numbered 1, 2, ...
+-- Total time on the drug can be calculated from this table by grouping using icustay_id
+
+-- Get drug administration data from CareVue first
+with vasocv1 as
+(
+ select
+ icustay_id, charttime
+ -- case statement determining whether the itemid is an instance of vasopressor usage
+ , max(case when itemid = 30051 then 1 else 0 end) as vaso -- vasopressin
+
+ -- the 'stopped' column indicates if a vasopressor has been disconnected
+ , max(case when itemid = 30051 and (stopped = 'Stopped' or stopped like 'D/C%') then 1
+ else 0 end) as vaso_stopped
+
+ , max(case when itemid = 30051 and rate is not null then 1 else 0 end) as vaso_null
+ , max(case when itemid = 30051 then rate else null end) as vaso_rate
+ , max(case when itemid = 30051 then amount else null end) as vaso_amount
+
+ from inputevents_cv
+ where itemid = 30051 -- vasopressin
+ group by icustay_id, charttime
+)
+
+, vasocv2 as
+(
+ select v.*
+ , sum(vaso_null) over (partition by icustay_id order by charttime) as vaso_partition
+ from
+ vasocv1 v
+)
+
+, vasocv3 as
+(
+ select v.*
+ , first_value(vaso_rate) over (partition by icustay_id, vaso_partition order by charttime) as vaso_prevrate_ifnull
+ from
+ vasocv2 v
+)
+
+, vasocv4 as
+(
+ select
+ icustay_id
+ , charttime
+ -- , (charttime - (lag(charttime, 1) over (partition by icustay_id, vaso order by charttime))) as delta
+
+ , vaso
+ , vaso_rate
+ , vaso_amount
+ , vaso_stopped
+ , vaso_prevrate_ifnull
+
+ -- We define start time here
+ , case
+ when vaso = 0 then null
+
+ -- if this is the first instance of the vasoactive drug
+ when vaso_rate > 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso, vaso_null
+ order by charttime
+ )
+ is null
+ then 1
+
+ -- you often get a string of 0s
+ -- we decide not to set these as 1, just because it makes vasonum sequential
+ when vaso_rate = 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 0
+ then 0
+
+ -- sometimes you get a string of null, associated with 0 volumes
+ -- same reason as before, we decide not to set these as 1
+ -- vaso_prevrate_ifnull is equal to the previous value *iff* the current value is null
+ when vaso_prevrate_ifnull = 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 0
+ then 0
+
+ -- If the last recorded rate was 0, newvaso = 1
+ when lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ ) = 0
+ then 1
+
+ -- If the last recorded vaso was D/C'd, newvaso = 1
+ when
+ lag(vaso_stopped,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ )
+ = 1 then 1
+
+ -- ** not sure if the below is needed
+ --when (charttime - (lag(charttime, 1) over (partition by icustay_id, vaso order by charttime))) > (interval '4 hours') then 1
+ else null
+ end as vaso_start
+ from
+ vasocv3
+)
+-- propagate start/stop flags forward in time
+
+, vasocv5 as
+(
+ select v.*
+ , sum(vaso_start) over (partition by icustay_id, vaso order by charttime) as vaso_first
+ from
+ vasocv4 v
+)
+
+, vasocv6 as
+(
+ select v.*
+ -- We define end time here
+ , case
+ when vaso = 0
+ then null
+
+ -- If the recorded vaso was D/C'd, this is an end time
+ when vaso_stopped = 1
+ then vaso_first
+
+ -- If the rate is zero, this is the end time
+ when vaso_rate = 0
+ then vaso_first
+
+ -- the last row in the table is always a potential end time
+ -- this captures patients who die/are discharged while on vasopressors
+ -- in principle, this could add an extra end time for the vasopressor
+ -- however, since we later group on vaso_start, any extra end times are ignored
+ when lead(charttime,1)
+ over
+ (
+ partition by icustay_id, vaso
+ order by charttime
+ ) is null
+ then vaso_first
+
+ else null
+ end as vaso_stop
+ from vasocv5 v
+)
+
+-- -- if you want to look at the results of the table before grouping:
+-- select
+-- icustay_id, charttime, vaso, vaso_rate, vaso_amount
+-- , case when vaso_stopped = 1 then 'Y' else '' end as stopped
+-- , vaso_start
+-- , vaso_first
+-- , vaso_stop
+-- from vasocv6 order by charttime;
+
+, vasocv as
+(
+ -- below groups together vasopressor administrations into groups
+ select
+ icustay_id
+ -- the first non-null rate is considered the starttime
+ , min(case when vaso_rate is not null then charttime else null end) as starttime
+ -- the *first* time the first/last flags agree is the stop time for this duration
+ , min(case when vaso_first = vaso_stop then charttime else null end) as endtime
+ from vasocv6
+ where
+ vaso_first is not null -- bogus data
+ and
+ vaso_first != 0 -- sometimes *only* a rate of 0 appears, i.e. the drug is never actually delivered
+ and
+ icustay_id is not null -- there are data for "floating" admissions, we don't worry about these
+ group by icustay_id, vaso_first
+ having -- ensure start time is not the same as end time
+ min(charttime) != min(case when vaso_first = vaso_stop then charttime else null end)
+ and
+ max(vaso_rate) > 0 -- if the rate was always 0 or null, we consider it not a real drug delivery
+)
+
+select
+ icustay_id
+ -- generate a sequential integer for convenience
+ , row_number() over (partition by icustay_id order by starttime) as vasonum
+ , starttime, endtime
+ , round((cast(extract(epoch from (endtime - starttime))/(60*60) as numeric)), 8) as duration_hours
+ -- add durations
+from
+ vasocv
+order by icustay_id, vasonum;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/13_vasopressor_durations.sql b/mimic-iii-carevue/code/concepts/13_vasopressor_durations.sql
new file mode 100644
index 000000000..0e0c84bfd
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/13_vasopressor_durations.sql
@@ -0,0 +1,263 @@
+drop table if exists vasopressor_durations; create table vasopressor_durations as
+-- This query extracts durations of vasopressor administration
+-- It groups together any administration of the below list of drugs:
+-- norepinephrine - 30047,30120
+-- epinephrine - 30044,30119,30309
+-- phenylephrine - 30127,30128
+-- vasopressin - 30051,(42273, 42802 also for 2 patients)
+-- dopamine - 30043,30307
+-- dobutamine - 30042,30306
+-- milrinone - 30125
+
+-- Consecutive administrations are numbered 1, 2, ...
+-- Total time on the drug can be calculated from this table
+-- by grouping using icustay_id
+
+-- select only the itemids from the inputevents_cv table related to vasopressors
+with io_cv as
+(
+ select
+ icustay_id, charttime, itemid, stopped
+ -- itemids (42273, 42802) accidentally store rate in amount column
+ , case
+ when itemid in (42273, 42802)
+ then amount
+ else rate
+ end as rate
+ , case
+ when itemid in (42273, 42802)
+ then rate
+ else amount
+ end as amount
+ from inputevents_cv
+ where itemid in
+ (
+ 30047,30120,30044,30119,30309,30127
+ , 30128,30051,30043,30307,30042,30306,30125
+ , 42273, 42802
+ )
+)
+
+, vasocv1 as
+(
+ select
+ icustay_id, charttime, itemid
+ -- case statement determining whether the ITEMID is an instance of vasopressor usage
+ , 1 as vaso
+
+ -- the 'stopped' column indicates if a vasopressor has been disconnected
+ , max(case when (stopped = 'Stopped' or stopped like 'D/C%') then 1
+ else 0 end) as vaso_stopped
+
+ , max(case when rate is not null then 1 else 0 end) as vaso_null
+ , max(rate) as vaso_rate
+ , max(amount) as vaso_amount
+ from io_cv
+ group by icustay_id, charttime, itemid
+)
+
+, vasocv2 as
+(
+ select v.*
+ , sum(vaso_null) over (partition by icustay_id, itemid order by charttime) as vaso_partition
+ from
+ vasocv1 v
+)
+
+, vasocv3 as
+(
+ select v.*
+ , first_value(vaso_rate) over (partition by icustay_id, itemid, vaso_partition order by charttime) as vaso_prevrate_ifnull
+ from
+ vasocv2 v
+)
+
+, vasocv4 as
+(
+ select
+ icustay_id
+ , charttime
+ , itemid
+ -- , (charttime - (lag(charttime, 1) over (partition by icustay_id, vaso order by charttime))) as delta
+
+ , vaso
+ , vaso_rate
+ , vaso_amount
+ , vaso_stopped
+ , vaso_prevrate_ifnull
+
+ -- We define start time here
+ , case
+ when vaso = 0 then null
+
+ -- if this is the first instance of the vasoactive drug
+ when vaso_rate > 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, itemid, vaso, vaso_null
+ order by charttime
+ )
+ is null
+ then 1
+
+ -- you often get a string of 0s
+ -- we decide not to set these as 1, just because it makes vasonum sequential
+ when vaso_rate = 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, itemid, vaso
+ order by charttime
+ )
+ = 0
+ then 0
+
+ -- sometimes you get a string of NULL, associated with 0 volumes
+ -- same reason as before, we decide not to set these as 1
+ -- vaso_prevrate_ifnull is equal to the previous value *iff* the current value is null
+ when vaso_prevrate_ifnull = 0 and
+ lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, itemid, vaso
+ order by charttime
+ )
+ = 0
+ then 0
+
+ -- If the last recorded rate was 0, newvaso = 1
+ when lag(vaso_prevrate_ifnull,1)
+ over
+ (
+ partition by icustay_id, itemid, vaso
+ order by charttime
+ ) = 0
+ then 1
+
+ -- If the last recorded vaso was D/C'd, newvaso = 1
+ when
+ lag(vaso_stopped,1)
+ over
+ (
+ partition by icustay_id, itemid, vaso
+ order by charttime
+ )
+ = 1 then 1
+
+ -- ** not sure if the below is needed
+ --when (charttime - (lag(charttime, 1) over (partition by icustay_id, vaso order by charttime))) > (interval '4 hours') then 1
+ else null
+ end as vaso_start
+ from
+ vasocv3
+)
+-- propagate start/stop flags forward in time
+
+, vasocv5 as
+(
+ select v.*
+ , sum(vaso_start) over (partition by icustay_id, itemid, vaso order by charttime) as vaso_first
+ from
+ vasocv4 v
+)
+
+, vasocv6 as
+(
+ select v.*
+ -- We define end time here
+ , case
+ when vaso = 0
+ then null
+
+ -- If the recorded vaso was D/C'd, this is an end time
+ when vaso_stopped = 1
+ then vaso_first
+
+ -- If the rate is zero, this is the end time
+ when vaso_rate = 0
+ then vaso_first
+
+ -- the last row in the table is always a potential end time
+ -- this captures patients who die/are discharged while on vasopressors
+ -- in principle, this could add an extra end time for the vasopressor
+ -- however, since we later group on vaso_start, any extra end times are ignored
+ when lead(charttime,1)
+ over
+ (
+ partition by icustay_id, itemid, vaso
+ order by charttime
+ ) is null
+ then vaso_first
+
+ else null
+ end as vaso_stop
+ from vasocv5 v
+)
+
+-- -- if you want to look at the results of the table before grouping:
+-- select
+-- icustay_id, charttime, vaso, vaso_rate, vaso_amount
+-- , case when vaso_stopped = 1 then 'Y' else '' end as stopped
+-- , vaso_start
+-- , vaso_first
+-- , vaso_stop
+-- from vasocv6 order by charttime;
+
+, vasocv as
+(
+ -- below groups together vasopressor administrations into groups
+ select
+ icustay_id
+ , itemid
+ -- the first non-null rate is considered the starttime
+ , min(case when vaso_rate is not null then charttime else null end) as starttime
+ -- the *first* time the first/last flags agree is the stop time for this duration
+ , min(case when vaso_first = vaso_stop then charttime else null end) as endtime
+ from vasocv6
+ where
+ vaso_first is not null -- bogus data
+ and
+ vaso_first != 0 -- sometimes *only* a rate of 0 appears, i.e. the drug is never actually delivered
+ and
+ icustay_id is not null -- there are data for "floating" admissions, we don't worry about these
+ group by icustay_id, itemid, vaso_first
+ having -- ensure start time is not the same as end time
+ min(charttime) != min(case when vaso_first = vaso_stop then charttime else null end)
+ and
+ max(vaso_rate) > 0 -- if the rate was always 0 or null, we consider it not a real drug delivery
+)
+-- we do not group by ITEMID in below query
+-- this is because we want to collapse all vasopressors together
+, vasocv_grp as
+(
+ select
+ s1.icustay_id,
+ s1.starttime,
+ min(t1.endtime) as endtime
+ from vasocv s1
+ inner join vasocv t1
+ on s1.icustay_id = t1.icustay_id
+ and s1.starttime <= t1.endtime
+ and not exists(select * from vasocv t2
+ where t1.icustay_id = t2.icustay_id
+ and t1.endtime >= t2.starttime
+ and t1.endtime < t2.endtime)
+ where not exists(select * from vasocv s2
+ where s1.icustay_id = s2.icustay_id
+ and s1.starttime > s2.starttime
+ and s1.starttime <= s2.endtime)
+ group by s1.icustay_id, s1.starttime
+ order by s1.icustay_id, s1.starttime
+)
+
+select
+ icustay_id
+ -- generate a sequential integer for convenience
+ , row_number() over (partition by icustay_id order by starttime) as vasonum
+ , starttime, endtime
+ , round((cast(extract(epoch from (endtime - starttime))/(60*60) as numeric)), 8) as duration_hours
+ -- add durations
+from
+ vasocv_grp
+order by icustay_id, vasonum;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/14_weight_durations.sql b/mimic-iii-carevue/code/concepts/14_weight_durations.sql
new file mode 100644
index 000000000..4fa05a7ee
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/14_weight_durations.sql
@@ -0,0 +1,187 @@
+drop table if exists weight_durations; create table weight_durations as
+
+with wt_neonate as
+(
+ select c.icustay_id, c.charttime
+ , max(case when c.itemid = 3580 then c.valuenum end) as wt_kg
+ , max(case when c.itemid = 3581 then c.valuenum end) as wt_lb
+ , max(case when c.itemid = 3582 then c.valuenum end) as wt_oz
+ from chartevents c
+ where c.itemid in (3580, 3581, 3582)
+ and c.icustay_id is not null
+ and coalesce(c.error, 0) = 0
+ -- wt_oz/wt_lb/wt_kg are only 0 erroneously, so drop these rows
+ and c.valuenum > 0
+ -- a separate query was run to manually verify only 1 value exists per
+ -- icustay_id/charttime/itemid grouping
+ -- therefore, we can use max() across itemid to collapse these values to 1 row per group
+ group by c.icustay_id, c.charttime
+)
+
+, birth_wt as
+(
+ select c.icustay_id, c.charttime
+ , max(
+ case
+ when c.itemid = 4183 then
+ -- clean free-text birth weight data
+ case
+ -- ignore value if there are any non-numeric characters
+ when c.value ~ '[^0-9\\.]' then null
+ -- convert grams to kd
+ when cast(c.value as numeric) > 100 then cast(c.value as numeric)/1000
+ -- keep kg as is, filtering bad values (largest baby ever born was conveniently 9.98kg)
+ when cast(c.value as numeric) < 10 then cast(c.value as numeric)
+ -- ignore other values (those between 10-100) - junk data
+ else null end
+ -- itemid 3723 happily has all numeric data - also doesn't store any grams data
+ when c.itemid = 3723 and c.valuenum < 10 then c.valuenum
+ else null end) as wt_kg
+ from chartevents c
+ where c.itemid in (3723, 4183)
+ and c.icustay_id is not null
+ and coalesce(c.error, 0) = 0
+ -- a separate query was run to manually verify only 1 value exists per
+ -- icustay_id/charttime/itemid grouping
+ -- therefore, we can use max() across itemid to collapse these values to 1 row per group
+ group by c.icustay_id, c.charttime
+)
+
+, wt_stg as
+(
+ select
+ c.icustay_id
+ , c.charttime
+ , case when c.itemid in (762,226512) then 'admit'
+ else 'daily' end as weight_type
+ -- TODO: eliminate obvious outliers if there is a reasonable weight
+ , c.valuenum as weight
+ from chartevents c
+ where c.valuenum is not null
+ and c.itemid in
+ (
+ 762,226512 -- Admit Wt
+ , 763,224639 -- Daily Weight
+ )
+ and c.icustay_id is not null
+ and c.valuenum > 0
+ -- exclude rows marked as error
+ and coalesce(c.error, 0) = 0
+ union all
+ select
+ n.icustay_id
+ , n.charttime
+ , 'daily' as weight_type
+ , case
+ when wt_kg is not null then wt_kg
+ when wt_lb is not null then wt_lb*0.45359237 + wt_oz*0.0283495231
+ else null end as weight
+ from wt_neonate n
+ union all
+ select
+ b.icustay_id
+ , b.charttime
+ -- birth weight of neonates is treated as admission weight
+ , 'admit' as weight_type
+ , wt_kg as weight
+ from birth_wt b
+)
+
+, wt_stg0 as
+(
+ select icustay_id, charttime, weight_type, weight
+ from wt_stg
+)
+
+-- assign ascending row number
+, wt_stg1 as
+(
+ select
+ icustay_id
+ , charttime
+ , weight_type
+ , weight
+ , row_number() over (partition by icustay_id, weight_type order by charttime) as rn
+ from wt_stg0
+ where weight is not null
+)
+
+-- change charttime to intime for the first admission weight recorded
+, wt_stg2 as
+(
+ select
+ wt_stg1.icustay_id
+ , ie.intime, ie.outtime
+ , case when wt_stg1.weight_type = 'admit' and wt_stg1.rn = 1
+ then ie.intime - interval '2 hour'
+ else wt_stg1.charttime end as starttime
+ , wt_stg1.weight
+ from wt_stg1
+ inner join icustays ie
+ on ie.icustay_id = wt_stg1.icustay_id
+)
+, wt_stg3 as
+(
+ select
+ icustay_id
+ , intime, outtime
+ , starttime
+ , coalesce(
+ lead(starttime) over (partition by icustay_id order by starttime),
+ greatest(outtime, starttime) + interval '2 hour'
+ ) as endtime
+ , weight
+ from wt_stg2
+)
+-- this table is the start/stop times from admit/daily weight in charted data
+, wt1 as
+(
+ select
+ icustay_id
+ , starttime
+ , coalesce(endtime,
+ lead(starttime) over (partition by icustay_id order by starttime),
+ -- impute ICU discharge as the end of the final weight measurement
+ -- plus a 2 hour "fuzziness" window
+ outtime + interval '2 hour'
+ ) as endtime
+ , weight
+ from wt_stg3
+)
+-- if the intime for the patient is < the first charted daily weight
+-- then we will have a "gap" at the start of their stay
+-- to prevent this, we look for these gaps and backfill the first weight
+, wt_fix as
+(
+ select ie.icustay_id
+ -- we add a 2 hour "fuzziness" window
+ , ie.intime - interval '2 hour' as starttime
+ , wt.starttime as endtime
+ , wt.weight
+ from icustays ie
+ inner join
+ -- the below subquery returns one row for each unique icustay_id
+ -- the row contains: the first starttime and the corresponding weight
+ (
+ select wt1.icustay_id, wt1.starttime, wt1.weight
+ , row_number() over (partition by wt1.icustay_id order by wt1.starttime) as rn
+ from wt1
+ ) wt
+ on ie.icustay_id = wt.icustay_id
+ and wt.rn = 1
+ and ie.intime < wt.starttime
+)
+-- add the backfill rows to the main weight table
+select
+ wt1.icustay_id
+ , wt1.starttime
+ , wt1.endtime
+ , wt1.weight
+from wt1
+union all
+select
+ wt_fix.icustay_id
+ , wt_fix.starttime
+ , wt_fix.endtime
+ , wt_fix.weight
+from wt_fix;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/15_elixhauser_ahrq_v37.sql b/mimic-iii-carevue/code/concepts/15_elixhauser_ahrq_v37.sql
new file mode 100644
index 000000000..fb43e4dec
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/15_elixhauser_ahrq_v37.sql
@@ -0,0 +1,991 @@
+drop table if exists elixhauser_ahrq_v37; create table elixhauser_ahrq_v37 as
+-- This code uses the latest version of Elixhauser provided by AHRQ
+
+with eliflg as
+(
+select hadm_id, seq_num, icd9_code
+-- note that these codes will seem incomplete at first
+-- for example, CHF is missing a lot of codes referenced in the literature (402.11, 402.91, etc)
+-- these codes are captured by hypertension flags instead
+-- later there are some complicated rules which confirm/reject those codes as chf
+, case
+ when icd9_code = '39891' then 1
+ when icd9_code between '4280' and '4289' then 1
+ end as chf /* Congestive heart failure */
+
+-- cardiac arrhythmias is removed in up to date versions
+, case
+ when icd9_code = '42610' then 1
+ when icd9_code = '42611' then 1
+ when icd9_code = '42613' then 1
+ when icd9_code between '4262' and '42653' then 1
+ when icd9_code between '4266' and '42689' then 1
+ when icd9_code = '4270' then 1
+ when icd9_code = '4272' then 1
+ when icd9_code = '42731' then 1
+ when icd9_code = '42760' then 1
+ when icd9_code = '4279' then 1
+ when icd9_code = '7850' then 1
+ when icd9_code between 'V450' and 'V4509' then 1
+ when icd9_code between 'V533' and 'V5339' then 1
+ end as arythm /* Cardiac arrhythmias */
+
+, case
+ when icd9_code between '09320' and '09324' then 1
+ when icd9_code between '3940' and '3971' then 1
+ when icd9_code = '3979' then 1
+ when icd9_code between '4240' and '42499' then 1
+ when icd9_code between '7463' and '7466' then 1
+ when icd9_code = 'V422' then 1
+ when icd9_code = 'V433' then 1
+ end as valve /* Valvular disease */
+
+, case
+ when icd9_code between '41511' and '41519' then 1
+ when icd9_code between '4160' and '4169' then 1
+ when icd9_code = '4179' then 1
+ end as pulmcirc /* Pulmonary circulation disorder */
+
+, case
+ when icd9_code between '4400' and '4409' then 1
+ when icd9_code between '44100' and '4419' then 1
+ when icd9_code between '4420' and '4429' then 1
+ when icd9_code between '4431' and '4439' then 1
+ when icd9_code between '44421' and '44422' then 1
+ when icd9_code = '4471' then 1
+ when icd9_code = '449' then 1
+ when icd9_code = '5571' then 1
+ when icd9_code = '5579' then 1
+ when icd9_code = 'V434' then 1
+ end as perivasc /* Peripheral vascular disorder */
+
+, case
+ when icd9_code = '4011' then 1
+ when icd9_code = '4019' then 1
+ when icd9_code between '64200' and '64204' then 1
+ end as htn /* Hypertension, uncomplicated */
+
+, case
+ when icd9_code = '4010' then 1
+ when icd9_code = '4372' then 1
+ end as htncx /* Hypertension, complicated */
+
+
+ /******************************************************************/
+ /* The following are special, temporary formats used in the */
+ /* creation of the hypertension complicated comorbidity when */
+ /* overlapping with congestive heart failure or renal failure */
+ /* occurs. These temporary formats are referenced in the program */
+ /* called comoanaly2009.txt. */
+ /******************************************************************/
+, case
+ when icd9_code between '64220' and '64224' then 1
+ end as htnpreg /* Pre-existing hypertension complicating pregnancy */
+
+, case
+ when icd9_code = '40200' then 1
+ when icd9_code = '40210' then 1
+ when icd9_code = '40290' then 1
+ when icd9_code = '40509' then 1
+ when icd9_code = '40519' then 1
+ when icd9_code = '40599' then 1
+ end as htnwochf /* Hypertensive heart disease without heart failure */
+
+, case
+ when icd9_code = '40201' then 1
+ when icd9_code = '40211' then 1
+ when icd9_code = '40291' then 1
+ end as htnwchf /* Hypertensive heart disease with heart failure */
+
+, case
+ when icd9_code = '40300' then 1
+ when icd9_code = '40310' then 1
+ when icd9_code = '40390' then 1
+ when icd9_code = '40501' then 1
+ when icd9_code = '40511' then 1
+ when icd9_code = '40591' then 1
+ when icd9_code between '64210' and '64214' then 1
+ end as hrenworf /* Hypertensive renal disease without renal failure */
+
+, case
+ when icd9_code = '40301' then 1
+ when icd9_code = '40311' then 1
+ when icd9_code = '40391' then 1
+ end as hrenwrf /* Hypertensive renal disease with renal failure */
+
+, case
+ when icd9_code = '40400' then 1
+ when icd9_code = '40410' then 1
+ when icd9_code = '40490' then 1
+ end as hhrwohrf /* Hypertensive heart and renal disease without heart or renal failure */
+
+, case
+ when icd9_code = '40401' then 1
+ when icd9_code = '40411' then 1
+ when icd9_code = '40491' then 1
+ end as hhrwchf /* Hypertensive heart and renal disease with heart failure */
+
+, case
+ when icd9_code = '40402' then 1
+ when icd9_code = '40412' then 1
+ when icd9_code = '40492' then 1
+ end as hhrwrf /* Hypertensive heart and renal disease with renal failure */
+
+, case
+ when icd9_code = '40403' then 1
+ when icd9_code = '40413' then 1
+ when icd9_code = '40493' then 1
+ end as hhrwhrf /* Hypertensive heart and renal disease with heart and renal failure */
+
+, case
+ when icd9_code between '64270' and '64274' then 1
+ when icd9_code between '64290' and '64294' then 1
+ end as ohtnpreg /* Other hypertension in pregnancy */
+
+ /******************** End Temporary Formats ***********************/
+
+, case
+ when icd9_code between '3420' and '3449' then 1
+ when icd9_code between '43820' and '43853' then 1
+ when icd9_code = '78072' then 1
+ end as para /* Paralysis */
+
+, case
+ when icd9_code between '3300' and '3319' then 1
+ when icd9_code = '3320' then 1
+ when icd9_code = '3334' then 1
+ when icd9_code = '3335' then 1
+ when icd9_code = '3337' then 1
+ when icd9_code in ('33371','33372','33379','33385','33394') then 1
+ when icd9_code between '3340' and '3359' then 1
+ when icd9_code = '3380' then 1
+ when icd9_code = '340' then 1
+ when icd9_code between '3411' and '3419' then 1
+ when icd9_code between '34500' and '34511' then 1
+ when icd9_code between '3452' and '3453' then 1
+ when icd9_code between '34540' and '34591' then 1
+ when icd9_code between '34700' and '34701' then 1
+ when icd9_code between '34710' and '34711' then 1
+ when icd9_code = '3483' then 1 -- discontinued icd-9
+ when icd9_code between '64940' and '64944' then 1
+ when icd9_code = '7687' then 1
+ when icd9_code between '76870' and '76873' then 1
+ when icd9_code = '7803' then 1
+ when icd9_code = '78031' then 1
+ when icd9_code = '78032' then 1
+ when icd9_code = '78033' then 1
+ when icd9_code = '78039' then 1
+ when icd9_code = '78097' then 1
+ when icd9_code = '7843' then 1
+ end as neuro /* Other neurological */
+
+, case
+ when icd9_code between '490' and '4928' then 1
+ when icd9_code between '49300' and '49392' then 1
+ when icd9_code between '494' and '4941' then 1
+ when icd9_code between '4950' and '505' then 1
+ when icd9_code = '5064' then 1
+ end as chrnlung /* Chronic pulmonary disease */
+
+, case
+ when icd9_code between '25000' and '25033' then 1
+ when icd9_code between '64800' and '64804' then 1
+ when icd9_code between '24900' and '24931' then 1
+ end as dm /* Diabetes w/o chronic complications*/
+
+, case
+ when icd9_code between '25040' and '25093' then 1
+ when icd9_code = '7751' then 1
+ when icd9_code between '24940' and '24991' then 1
+ end as dmcx /* Diabetes w/ chronic complications */
+
+, case
+ when icd9_code between '243' and '2442' then 1
+ when icd9_code = '2448' then 1
+ when icd9_code = '2449' then 1
+ end as hypothy /* Hypothyroidism */
+
+, case
+ when icd9_code = '585' then 1 -- discontinued code
+ when icd9_code = '5853' then 1
+ when icd9_code = '5854' then 1
+ when icd9_code = '5855' then 1
+ when icd9_code = '5856' then 1
+ when icd9_code = '5859' then 1
+ when icd9_code = '586' then 1
+ when icd9_code = 'V420' then 1
+ when icd9_code = 'V451' then 1
+ when icd9_code between 'V560' and 'V5632' then 1
+ when icd9_code = 'V568' then 1
+ when icd9_code between 'V4511' and 'V4512' then 1
+ end as renlfail /* Renal failure */
+
+, case
+ when icd9_code = '07022' then 1
+ when icd9_code = '07023' then 1
+ when icd9_code = '07032' then 1
+ when icd9_code = '07033' then 1
+ when icd9_code = '07044' then 1
+ when icd9_code = '07054' then 1
+ when icd9_code = '4560' then 1
+ when icd9_code = '4561' then 1
+ when icd9_code = '45620' then 1
+ when icd9_code = '45621' then 1
+ when icd9_code = '5710' then 1
+ when icd9_code = '5712' then 1
+ when icd9_code = '5713' then 1
+ when icd9_code between '57140' and '57149' then 1
+ when icd9_code = '5715' then 1
+ when icd9_code = '5716' then 1
+ when icd9_code = '5718' then 1
+ when icd9_code = '5719' then 1
+ when icd9_code = '5723' then 1
+ when icd9_code = '5728' then 1
+ when icd9_code = '5735' then 1
+ when icd9_code = 'V427' then 1
+ end as liver /* Liver disease */
+
+, case
+ when icd9_code = '53141' then 1
+ when icd9_code = '53151' then 1
+ when icd9_code = '53161' then 1
+ when icd9_code = '53170' then 1
+ when icd9_code = '53171' then 1
+ when icd9_code = '53191' then 1
+ when icd9_code = '53241' then 1
+ when icd9_code = '53251' then 1
+ when icd9_code = '53261' then 1
+ when icd9_code = '53270' then 1
+ when icd9_code = '53271' then 1
+ when icd9_code = '53291' then 1
+ when icd9_code = '53341' then 1
+ when icd9_code = '53351' then 1
+ when icd9_code = '53361' then 1
+ when icd9_code = '53370' then 1
+ when icd9_code = '53371' then 1
+ when icd9_code = '53391' then 1
+ when icd9_code = '53441' then 1
+ when icd9_code = '53451' then 1
+ when icd9_code = '53461' then 1
+ when icd9_code = '53470' then 1
+ when icd9_code = '53471' then 1
+ when icd9_code = '53491' then 1
+ end as ulcer /* Chronic Peptic ulcer disease (includes bleeding only if obstruction is also present) */
+
+, case
+ when icd9_code between '042' and '0449' then 1
+ end as aids /* HIV and AIDS */
+
+, case
+ when icd9_code between '20000' and '20238' then 1
+ when icd9_code between '20250' and '20301' then 1
+ when icd9_code = '2386' then 1
+ when icd9_code = '2733' then 1
+ when icd9_code between '20302' and '20382' then 1
+ end as lymph /* Lymphoma */
+
+, case
+ when icd9_code between '1960' and '1991' then 1
+ when icd9_code between '20970' and '20975' then 1
+ when icd9_code = '20979' then 1
+ when icd9_code = '78951' then 1
+ end as mets /* Metastatic cancer */
+
+, case
+ when icd9_code between '1400' and '1729' then 1
+ when icd9_code between '1740' and '1759' then 1
+ when icd9_code between '179' and '1958' then 1
+ when icd9_code between '20900' and '20924' then 1
+ when icd9_code between '20925' and '2093' then 1
+ when icd9_code between '20930' and '20936' then 1
+ when icd9_code between '25801' and '25803' then 1
+ end as tumor /* Solid tumor without metastasis */
+
+, case
+ when icd9_code = '7010' then 1
+ when icd9_code between '7100' and '7109' then 1
+ when icd9_code between '7140' and '7149' then 1
+ when icd9_code between '7200' and '7209' then 1
+ when icd9_code = '725' then 1
+ end as arth /* Rheumatoid arthritis/collagen vascular diseases */
+
+, case
+ when icd9_code between '2860' and '2869' then 1
+ when icd9_code = '2871' then 1
+ when icd9_code between '2873' and '2875' then 1
+ when icd9_code between '64930' and '64934' then 1
+ when icd9_code = '28984' then 1
+ end as coag /* Coagulation deficiency */
+
+, case
+ when icd9_code = '2780' then 1
+ when icd9_code = '27800' then 1
+ when icd9_code = '27801' then 1
+ when icd9_code = '27803' then 1
+ when icd9_code between '64910' and '64914' then 1
+ when icd9_code between 'V8530' and 'V8539' then 1
+ when icd9_code = 'V854' then 1 -- hierarchy used for AHRQ v3.6 and earlier
+ when icd9_code between 'V8541' and 'V8545' then 1
+ when icd9_code = 'V8554' then 1
+ when icd9_code = '79391' then 1
+ end as obese /* Obesity */
+
+, case
+ when icd9_code between '260' and '2639' then 1
+ when icd9_code between '78321' and '78322' then 1
+ end as wghtloss /* Weight loss */
+
+, case
+ when icd9_code between '2760' and '2769' then 1
+ end as lytes /* Fluid and electrolyte disorders - note:
+ this comorbidity should be dropped when
+ used with the AHRQ Patient Safety Indicators*/
+, case
+ when icd9_code = '2800' then 1
+ when icd9_code between '64820' and '64824' then 1
+ end as bldloss /* Blood loss anemia */
+
+, case
+ when icd9_code between '2801' and '2819' then 1
+ when icd9_code between '28521' and '28529' then 1
+ when icd9_code = '2859' then 1
+ end as anemdef /* Deficiency anemias */
+
+, case
+ when icd9_code between '2910' and '2913' then 1
+ when icd9_code = '2915' then 1
+ when icd9_code = '2918' then 1
+ when icd9_code = '29181' then 1
+ when icd9_code = '29182' then 1
+ when icd9_code = '29189' then 1
+ when icd9_code = '2919' then 1
+ when icd9_code between '30300' and '30393' then 1
+ when icd9_code between '30500' and '30503' then 1
+ end as alcohol /* Alcohol abuse */
+
+, case
+ when icd9_code = '2920' then 1
+ when icd9_code between '29282' and '29289' then 1
+ when icd9_code = '2929' then 1
+ when icd9_code between '30400' and '30493' then 1
+ when icd9_code between '30520' and '30593' then 1
+ when icd9_code between '64830' and '64834' then 1
+ end as drug /* Drug abuse */
+
+, case
+ when icd9_code between '29500' and '2989' then 1
+ when icd9_code = '29910' then 1
+ when icd9_code = '29911' then 1
+ end as psych /* Psychoses */
+
+, case
+ when icd9_code = '3004' then 1
+ when icd9_code = '30112' then 1
+ when icd9_code = '3090' then 1
+ when icd9_code = '3091' then 1
+ when icd9_code = '311' then 1
+ end as depress /* Depression */
+from diagnoses_icd icd
+where seq_num = 1
+)
+-- collapse the icd9_code specific flags into hadm_id specific flags
+-- this groups comorbidities together for a single patient admission
+, eligrp as
+(
+ select hadm_id
+ , max(chf) as chf
+ , max(arythm) as arythm
+ , max(valve) as valve
+ , max(pulmcirc) as pulmcirc
+ , max(perivasc) as perivasc
+ , max(htn) as htn
+ , max(htncx) as htncx
+ , max(htnpreg) as htnpreg
+ , max(htnwochf) as htnwochf
+ , max(htnwchf) as htnwchf
+ , max(hrenworf) as hrenworf
+ , max(hrenwrf) as hrenwrf
+ , max(hhrwohrf) as hhrwohrf
+ , max(hhrwchf) as hhrwchf
+ , max(hhrwrf) as hhrwrf
+ , max(hhrwhrf) as hhrwhrf
+ , max(ohtnpreg) as ohtnpreg
+ , max(para) as para
+ , max(neuro) as neuro
+ , max(chrnlung) as chrnlung
+ , max(dm) as dm
+ , max(dmcx) as dmcx
+ , max(hypothy) as hypothy
+ , max(renlfail) as renlfail
+ , max(liver) as liver
+ , max(ulcer) as ulcer
+ , max(aids) as aids
+ , max(lymph) as lymph
+ , max(mets) as mets
+ , max(tumor) as tumor
+ , max(arth) as arth
+ , max(coag) as coag
+ , max(obese) as obese
+ , max(wghtloss) as wghtloss
+ , max(lytes) as lytes
+ , max(bldloss) as bldloss
+ , max(anemdef) as anemdef
+ , max(alcohol) as alcohol
+ , max(drug) as drug
+ , max(psych) as psych
+ , max(depress) as depress
+from eliflg
+group by hadm_id
+)
+
+-- DRG FILTER --
+, msdrg as
+(
+select
+ hadm_id
+/**** V29 MS-DRG Formats ****/
+
+/* Cardiac */
+, case
+ when d.drg_code between 001 and 002 then 1
+ when d.drg_code between 215 and 238 then 1
+ when d.drg_code between 242 and 252 then 1
+ when d.drg_code between 253 and 254 then 1
+ when d.drg_code between 258 and 262 then 1
+ when d.drg_code between 265 and 267 then 1
+ when d.drg_code between 280 and 293 then 1
+ when d.drg_code between 296 and 298 then 1
+ when d.drg_code between 302 and 303 then 1
+ when d.drg_code between 306 and 313 then 1
+else 0 end as carddrg
+
+/* Peripheral vascular */
+, case
+ when d.drg_code between 299 and 301 then 1
+else 0 end as peridrg
+
+/* Renal */
+, case
+ when d.drg_code = 652 then 1
+ when d.drg_code between 656 and 661 then 1
+ when d.drg_code between 673 and 675 then 1
+ when d.drg_code between 682 and 700 then 1
+else 0 end as renaldrg
+
+/* Nervous system */
+, case
+ when d.drg_code between 020 and 042 then 1
+ when d.drg_code between 052 and 103 then 1
+else 0 end as nervdrg
+
+/* Cerebrovascular */
+, case
+ when d.drg_code between 020 and 022 then 1
+ when d.drg_code between 034 and 039 then 1
+ when d.drg_code between 064 and 072 then 1
+else 0 end as ceredrg
+
+/* COPD asthma */
+, case
+ when d.drg_code between 190 and 192 then 1
+ when d.drg_code between 202 and 203 then 1
+else 0 end as pulmdrg
+
+/* Diabetes */
+, case
+ when d.drg_code between 637 and 639 then 1
+else 0 end as DIABDRG
+
+/* Thyroid endocrine */
+, case
+ when d.drg_code between 625 and 627 then 1
+ when d.drg_code between 643 and 645 then 1
+else 0 end as hypodrg
+
+/* Kidney transp, renal fail/dialysis */
+, case
+ when d.drg_code = 652 then 1
+ when d.drg_code between 682 and 685 then 1
+else 0 end as renfdrg
+
+/* Liver */
+, case
+ when d.drg_code between 420 and 425 then 1
+ when d.drg_code between 432 and 434 then 1
+ when d.drg_code between 441 and 446 then 1
+else 0 end as liverdrg
+
+/* GI hemorrhage or ulcer */
+, case
+ when d.drg_code between 377 and 384 then 1
+else 0 end as ulcedrg
+
+/* Human immunodeficiency virus */
+, case
+ when d.drg_code between 969 and 970 then 1
+ when d.drg_code between 974 and 977 then 1
+else 0 end as hivdrg
+
+/* Leukemia/lymphoma */
+, case
+ when d.drg_code between 820 and 830 then 1
+ when d.drg_code between 834 and 849 then 1
+else 0 end as leukdrg
+
+/* Cancer, lymphoma */
+, case
+ when d.drg_code = 054 then 1
+ when d.drg_code = 055 then 1
+ when d.drg_code between 146 and 148 then 1
+ when d.drg_code between 180 and 182 then 1
+ when d.drg_code between 374 and 376 then 1
+ when d.drg_code between 435 and 437 then 1
+ when d.drg_code between 542 and 544 then 1
+ when d.drg_code between 582 and 585 then 1
+ when d.drg_code between 597 and 599 then 1
+ when d.drg_code between 656 and 658 then 1
+ when d.drg_code between 686 and 688 then 1
+ when d.drg_code between 715 and 716 then 1
+ when d.drg_code between 722 and 724 then 1
+ when d.drg_code between 736 and 741 then 1
+ when d.drg_code between 754 and 756 then 1
+ when d.drg_code between 826 and 830 then 1
+ when d.drg_code between 843 and 849 then 1
+else 0 end as cancdrg
+
+/* Connective tissue */
+, case
+ when d.drg_code between 545 and 547 then 1
+else 0 end as arthdrg
+
+/* Nutrition/metabolic */
+, case
+ when d.drg_code between 640 and 641 then 1
+else 0 end as nutrdrg
+
+/* Anemia */
+, case
+ when d.drg_code between 808 and 812 then 1
+else 0 end as anemdrg
+
+/* Alcohol drug */
+, case
+ when d.drg_code between 894 and 897 then 1
+else 0 end as alcdrg
+
+/*Coagulation disorders*/
+, case
+ when d.drg_code = 813 then 1
+else 0 end as coagdrg
+
+/*Hypertensive Complicated */
+, case
+ when d.drg_code = 077 then 1
+ when d.drg_code = 078 then 1
+ when d.drg_code = 304 then 1
+else 0 end as htncxdrg
+
+/*Hypertensive Uncomplicated */
+, case
+ when d.drg_code = 079 then 1
+ when d.drg_code = 305 then 1
+else 0 end as htndrg
+
+/* Psychoses */
+, case
+ when d.drg_code = 885 then 1
+else 0 end as psydrg
+
+/* Obesity */
+, case
+ when d.drg_code between 619 and 621 then 1
+else 0 end as obesedrg
+
+/* Depressive Neuroses */
+, case
+ when d.drg_code = 881 then 1
+else 0 end as deprsdrg
+
+from
+(
+ select hadm_id, drg_type, cast(drg_code as numeric) as drg_code
+ from drgcodes
+ where drg_type = 'MS'
+) d
+
+)
+, hcfadrg as
+(
+select
+ hadm_id
+
+ /** V24 DRG Formats **/
+
+ /* Cardiac */
+ , case
+ when d.drg_code between 103 and 112 then 1
+ when d.drg_code between 115 and 118 then 1
+ when d.drg_code between 121 and 127 then 1
+ when d.drg_code = 129 then 1
+ when d.drg_code = 132 then 1
+ when d.drg_code = 133 then 1
+ when d.drg_code between 135 and 143 then 1
+ when d.drg_code between 514 and 518 then 1
+ when d.drg_code between 525 and 527 then 1
+ when d.drg_code between 535 and 536 then 1
+ when d.drg_code between 547 and 550 then 1
+ when d.drg_code between 551 and 558 then 1
+ else 0 end as carddrg
+
+ /* Peripheral vascular */
+ , case
+ when d.drg_code = 130 then 1
+ when d.drg_code = 131 then 1
+ else 0 end as peridrg
+
+ /* Renal */
+ , case
+ when d.drg_code between 302 and 305 then 1
+ when d.drg_code between 315 and 333 then 1
+
+ else 0 end as renaldrg
+
+ /* Nervous system */
+ , case
+ when d.drg_code between 1 and 35 then 1
+ when d.drg_code = 524 then 1
+ when d.drg_code between 528 and 534 then 1
+ when d.drg_code = 543 then 1
+ when d.drg_code between 559 and 564 then 1
+ when d.drg_code = 577 then 1
+
+ else 0 end as nervdrg
+
+ /* Cerebrovascular */
+ , case
+ when d.drg_code = 5 then 1
+ when d.drg_code between 14 and 17 then 1
+ when d.drg_code = 524 then 1
+ when d.drg_code = 528 then 1
+ when d.drg_code between 533 and 534 then 1
+ when d.drg_code = 577 then 1
+ else 0 end as ceredrg
+
+ /* COPD asthma */
+ , case
+ when d.drg_code = 88 then 1
+ when d.drg_code between 96 and 98 then 1
+
+ else 0 end as pulmdrg
+
+ /* Diabetes */
+ , case
+ when d.drg_code = 294 then 1
+ when d.drg_code = 295 then 1
+ else 0 end as diabdrg
+
+ /* Thyroid endocrine */
+ , case
+ when d.drg_code = 290 then 1
+ when d.drg_code = 300 then 1
+ when d.drg_code = 301 then 1
+
+ else 0 end as hypodrg
+
+ /* Kidney transp, renal fail/dialysis */
+ , case
+ when d.drg_code = 302 then 1
+ when d.drg_code = 316 then 1
+ when d.drg_code = 317 then 1
+ else 0 end as renfdrg
+
+ /* Liver */
+ , case
+ when d.drg_code between 199 and 202 then 1
+ when d.drg_code between 205 and 208 then 1
+
+ else 0 end as liverdrg
+
+ /* GI hemorrhage or ulcer */
+ , case
+ when d.drg_code between 174 and 178 then 1
+ else 0 end as ulcedrg
+
+ /* Human immunodeficiency virus */
+ , case
+ when d.drg_code = 488 then 1
+ when d.drg_code = 489 then 1
+ when d.drg_code = 490 then 1
+
+ else 0 end as hivdrg
+
+ /* Leukemia/lymphoma */
+ , case
+ when d.drg_code between 400 and 414 then 1
+ when d.drg_code = 473 then 1
+ when d.drg_code = 492 then 1
+ when d.drg_code between 539 and 540 then 1
+
+ else 0 end as leukdrg
+
+ /* Cancer, lymphoma */
+ , case
+ when d.drg_code = 10 then 1
+ when d.drg_code = 11 then 1
+ when d.drg_code = 64 then 1
+ when d.drg_code = 82 then 1
+ when d.drg_code = 172 then 1
+ when d.drg_code = 173 then 1
+ when d.drg_code = 199 then 1
+ when d.drg_code = 203 then 1
+ when d.drg_code = 239 then 1
+
+ when d.drg_code between 257 and 260 then 1
+ when d.drg_code = 274 then 1
+ when d.drg_code = 275 then 1
+ when d.drg_code = 303 then 1
+ when d.drg_code = 318 then 1
+ when d.drg_code = 319 then 1
+
+ when d.drg_code = 338 then 1
+ when d.drg_code = 344 then 1
+ when d.drg_code = 346 then 1
+ when d.drg_code = 347 then 1
+ when d.drg_code = 354 then 1
+ when d.drg_code = 355 then 1
+ when d.drg_code = 357 then 1
+ when d.drg_code = 363 then 1
+ when d.drg_code = 366 then 1
+
+ when d.drg_code = 367 then 1
+ when d.drg_code between 406 and 414 then 1
+ else 0 end as cancdrg
+
+ /* Connective tissue */
+ , case
+ when d.drg_code = 240 then 1
+ when d.drg_code = 241 then 1
+ else 0 end as arthdrg
+
+ /* Nutrition/metabolic */
+ , case
+ when d.drg_code between 296 and 298 then 1
+ else 0 end as nutrdrg
+
+ /* Anemia */
+ , case
+ when d.drg_code = 395 then 1
+ when d.drg_code = 396 then 1
+ when d.drg_code = 574 then 1
+ else 0 end as anemdrg
+
+ /* Alcohol drug */
+ , case
+ when d.drg_code between 433 and 437 then 1
+ when d.drg_code between 521 and 523 then 1
+ else 0 end as alcdrg
+
+ /* Coagulation disorders */
+ , case
+ when d.drg_code = 397 then 1
+ else 0 end as coagdrg
+
+ /* Hypertensive Complicated */
+ , case
+ when d.drg_code = 22 then 1
+ when d.drg_code = 134 then 1
+ else 0 end as htncxdrg
+
+ /* Hypertensive Uncomplicated */
+ , case
+ when d.drg_code = 134 then 1
+ else 0 end as htndrg
+
+ /* Psychoses */
+ , case
+ when d.drg_code = 430 then 1
+ else 0 end as psydrg
+
+ /* Obesity */
+ , case
+ when d.drg_code = 288 then 1
+ else 0 end as obesedrg
+
+ /* Depressive Neuroses */
+ , case
+ when d.drg_code = 426 then 1
+ else 0 end as deprsdrg
+
+ from
+ (
+ select hadm_id, drg_type, cast(drg_code as numeric) as drg_code
+ from drgcodes
+ where drg_type = 'HCFA'
+ ) d
+)
+-- merge DRG groups together
+, drggrp as
+(
+ select hadm_id
+, max(carddrg) as carddrg
+, max(peridrg) as peridrg
+, max(renaldrg) as renaldrg
+, max(nervdrg) as nervdrg
+, max(ceredrg) as ceredrg
+, max(pulmdrg) as pulmdrg
+, max(diabdrg) as diabdrg
+, max(hypodrg) as hypodrg
+, max(renfdrg) as renfdrg
+, max(liverdrg) as liverdrg
+, max(ulcedrg) as ulcedrg
+, max(hivdrg) as hivdrg
+, max(leukdrg) as leukdrg
+, max(cancdrg) as cancdrg
+, max(arthdrg) as arthdrg
+, max(nutrdrg) as nutrdrg
+, max(anemdrg) as anemdrg
+, max(alcdrg) as alcdrg
+, max(coagdrg) as coagdrg
+, max(htncxdrg) as htncxdrg
+, max(htndrg) as htndrg
+, max(psydrg) as psydrg
+, max(obesedrg) as obesedrg
+, max(deprsdrg) as deprsdrg
+from
+(
+ select d1.* from msdrg d1
+ union distinct
+ select d1.* from hcfadrg d1
+) d
+group by d.hadm_id
+)
+-- now merge these flags together to define elixhauser
+-- most are straightforward.. but hypertension flags are a bit more complicated
+select adm.subject_id, adm.hadm_id
+, case
+ when carddrg = 1 then 0 -- DRG filter
+
+ when chf = 1 then 1
+ when htnwchf = 1 then 1
+ when hhrwchf = 1 then 1
+ when hhrwhrf = 1 then 1
+ else 0 end as congestive_heart_failure
+, case
+ when carddrg = 1 then 0 -- DRG filter
+ when arythm = 1 then 1
+ else 0 end as cardiac_arrhythmias
+, case
+ when carddrg = 1 then 0
+ when valve = 1 then 1
+ else 0 end as valvular_disease
+, case
+ when carddrg = 1 or pulmdrg = 1 then 0
+ when pulmcirc = 1 then 1
+ else 0 end as pulmonary_circulation
+, case
+ when peridrg = 1 then 0
+ when perivasc = 1 then 1
+ else 0 end as peripheral_vascular
+
+-- we combine 'htn' and 'htncx' into 'HYPERTENSION'
+-- note 'htn' (hypertension) is only 1 if 'htncx' (complicated hypertension) is 0
+-- also if htncxdrg = 1, then htndrg = 1
+
+-- In the original Sas code, it appears that:
+-- HTN can be 1
+-- HTNCX is set to 0 by DRGs
+-- but HTN_C is still 1, because HTN is 1
+-- so we have to do this complex addition.
+,
+case
+ when
+(
+-- first hypertension
+case
+ when htndrg = 0 then 0
+ when htn = 1 then 1
+else 0 end
+)
++
+(
+-- next complicated hypertension
+case
+ when htncx = 1 and htncxdrg = 1 then 0
+
+ when htnpreg = 1 and htncxdrg = 1 then 0
+ when htnwochf = 1 and (htncxdrg = 1 OR carddrg = 1) then 0
+ when htnwchf = 1 and htncxdrg = 1 then 0
+ when htnwchf = 1 and carddrg = 1 then 0
+ when hrenworf = 1 and (htncxdrg = 1 or renaldrg = 1) then 0
+ when hrenwrf = 1 and htncxdrg = 1 then 0
+ when hrenwrf = 1 and renaldrg = 1 then 0
+ when hhrwohrf = 1 and (htncxdrg = 1 or carddrg = 1 or renaldrg = 1) then 0
+ when hhrwchf = 1 and (htncxdrg = 1 or carddrg = 1 or renaldrg = 1) then 0
+ when hhrwrf = 1 and (htncxdrg = 1 or carddrg = 1 or renaldrg = 1) then 0
+ when hhrwhrf = 1 and (htncxdrg = 1 or carddrg = 1 or renaldrg = 1) then 0
+ when ohtnpreg = 1 and (htncxdrg = 1 or carddrg = 1 or renaldrg = 1) then 0
+
+ when htncx = 1 then 1
+ when htnpreg = 1 then 1
+ when htnwochf = 1 then 1
+ when htnwchf = 1 then 1
+ when hrenworf = 1 then 1
+ when hrenwrf = 1 then 1
+ when hhrwohrf = 1 then 1
+ when hhrwchf = 1 then 1
+ when hhrwrf = 1 then 1
+ when hhrwhrf = 1 then 1
+ when ohtnpreg = 1 then 1
+ else 0 end
+)
+ > 0 then 1 else 0 end as hypertension
+
+, case when ceredrg = 1 then 0 when para = 1 then 1 else 0 end as paralysis
+, case when nervdrg = 1 then 0 when neuro = 1 then 1 else 0 end as other_neurological
+, case when pulmdrg = 1 then 0 when chrnlung = 1 then 1 else 0 end as chronic_pulmonary
+, case
+ -- only the more severe comorbidity (complicated diabetes) is kept
+ when diabdrg = 1 then 0
+ when dmcx = 1 then 0
+ when dm = 1 then 1
+ else 0 end as diabetes_uncomplicated
+, case when diabdrg = 1 then 0 when dmcx = 1 then 1 else 0 end as diabetes_complicated
+, case when hypodrg = 1 then 0 when hypothy = 1 then 1 else 0 end as hypothyroidism
+, case
+ when renaldrg = 1 then 0
+ when renlfail = 1 then 1
+ when hrenwrf = 1 then 1
+ when hhrwrf = 1 then 1
+ when hhrwhrf = 1 then 1
+ else 0 end as renal_failure
+
+, case when liverdrg = 1 then 0 when liver = 1 then 1 else 0 end as liver_disease
+, case when ulcedrg = 1 then 0 when ulcer = 1 then 1 else 0 end as peptic_ulcer
+, case when hivdrg = 1 then 0 when aids = 1 then 1 else 0 end as aids
+, case when leukdrg = 1 then 0 when lymph = 1 then 1 else 0 end as lymphoma
+, case when cancdrg = 1 then 0 when mets = 1 then 1 else 0 end as metastatic_cancer
+, case
+ when cancdrg = 1 then 0
+ -- only the more severe comorbidity (metastatic cancer) is kept
+ when mets = 1 then 0
+ when tumor = 1 then 1
+ else 0 end as solid_tumor
+, case when arthdrg = 1 then 0 when arth = 1 then 1 else 0 end as rheumatoid_arthritis
+, case when coagdrg = 1 then 0 when coag = 1 then 1 else 0 end as coagulopathy
+, case when nutrdrg = 1
+ OR obesedrg = 1 then 0 when obese = 1 then 1 else 0 end as obesity
+, case when nutrdrg = 1 then 0 when wghtloss = 1 then 1 else 0 end as weight_loss
+, case when nutrdrg = 1 then 0 when lytes = 1 then 1 else 0 end as fluid_electrolyte
+, case when anemdrg = 1 then 0 when bldloss = 1 then 1 else 0 end as blood_loss_anemia
+, case when anemdrg = 1 then 0 when anemdef = 1 then 1 else 0 end as deficiency_anemias
+, case when alcdrg = 1 then 0 when alcohol = 1 then 1 else 0 end as alcohol_abuse
+, case when alcdrg = 1 then 0 when drug = 1 then 1 else 0 end as drug_abuse
+, case when psydrg = 1 then 0 when psych = 1 then 1 else 0 end as psychoses
+, case when deprsdrg = 1 then 0 when depress = 1 then 1 else 0 end as depression
+
+from admissions adm
+left join eligrp eli
+ on adm.hadm_id = eli.hadm_id
+left join drggrp d
+ on adm.hadm_id = d.hadm_id
+order by adm.hadm_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/16_elixhauser_ahrq_v37_no_drg.sql b/mimic-iii-carevue/code/concepts/16_elixhauser_ahrq_v37_no_drg.sql
new file mode 100644
index 000000000..204921922
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/16_elixhauser_ahrq_v37_no_drg.sql
@@ -0,0 +1,522 @@
+drop table if exists elixhauser_ahrq_v37_no_drg; create table elixhauser_ahrq_v37_no_drg as
+-- This code uses the latest version of Elixhauser provided by AHRQ
+-- However, it does *not* filter based on diagnosis related groups (DRGs)
+-- As such, "comorbidities" identified are more likely to be associated with the primary reason for their hospital stay
+
+-- The code:
+-- removes "primary" ICD9_CODE (seq_num != 1)
+-- uses AHRQ published rules to define comorbidities
+with
+eliflg as
+(
+select hadm_id, seq_num, icd9_code
+-- note that these codes will seem incomplete at first
+-- for example, CHF is missing a lot of codes referenced in the literature (402.11, 402.91, etc)
+-- these codes are captured by hypertension flags instead
+-- later there are some complicated rules which confirm/reject those codes as chf
+, case
+ when icd9_code = '39891' then 1
+ when icd9_code between '4280' and '4289' then 1
+ end as chf /* Congestive heart failure */
+
+-- cardiac arrhythmias is removed in up to date versions
+, case
+ when icd9_code = '42610' then 1
+ when icd9_code = '42611' then 1
+ when icd9_code = '42613' then 1
+ when icd9_code between '4262' and '42653' then 1
+ when icd9_code between '4266' and '42689' then 1
+ when icd9_code = '4270' then 1
+ when icd9_code = '4272' then 1
+ when icd9_code = '42731' then 1
+ when icd9_code = '42760' then 1
+ when icd9_code = '4279' then 1
+ when icd9_code = '7850' then 1
+ when icd9_code between 'V450' and 'V4509' then 1
+ when icd9_code between 'V533' and 'V5339' then 1
+ end as arythm /* Cardiac arrhythmias */
+
+, case
+ when icd9_code between '09320' and '09324' then 1
+ when icd9_code between '3940' and '3971' then 1
+ when icd9_code = '3979' then 1
+ when icd9_code between '4240' and '42499' then 1
+ when icd9_code between '7463' and '7466' then 1
+ when icd9_code = 'V422' then 1
+ when icd9_code = 'V433' then 1
+ end as valve /* Valvular disease */
+
+, case
+ when icd9_code between '41511' and '41519' then 1
+ when icd9_code between '4160' and '4169' then 1
+ when icd9_code = '4179' then 1
+ end as pulmcirc /* Pulmonary circulation disorder */
+
+, case
+ when icd9_code between '4400' and '4409' then 1
+ when icd9_code between '44100' and '4419' then 1
+ when icd9_code between '4420' and '4429' then 1
+ when icd9_code between '4431' and '4439' then 1
+ when icd9_code between '44421' and '44422' then 1
+ when icd9_code = '4471' then 1
+ when icd9_code = '449' then 1
+ when icd9_code = '5571' then 1
+ when icd9_code = '5579' then 1
+ when icd9_code = 'V434' then 1
+ end as perivasc /* Peripheral vascular disorder */
+
+, case
+ when icd9_code = '4011' then 1
+ when icd9_code = '4019' then 1
+ when icd9_code between '64200' and '64204' then 1
+ end as htn /* Hypertension, uncomplicated */
+
+, case
+ when icd9_code = '4010' then 1
+ when icd9_code = '4372' then 1
+ end as htncx /* Hypertension, complicated */
+
+
+ /******************************************************************/
+ /* The following are special, temporary formats used in the */
+ /* creation of the hypertension complicated comorbidity when */
+ /* overlapping with congestive heart failure or renal failure */
+ /* occurs. These temporary formats are referenced in the program */
+ /* called comoanaly2009.txt. */
+ /******************************************************************/
+, case
+ when icd9_code between '64220' and '64224' then 1
+ end as htnpreg /* Pre-existing hypertension complicating pregnancy */
+
+, case
+ when icd9_code = '40200' then 1
+ when icd9_code = '40210' then 1
+ when icd9_code = '40290' then 1
+ when icd9_code = '40509' then 1
+ when icd9_code = '40519' then 1
+ when icd9_code = '40599' then 1
+ end as htnwochf /* Hypertensive heart disease without heart failure */
+
+, case
+ when icd9_code = '40201' then 1
+ when icd9_code = '40211' then 1
+ when icd9_code = '40291' then 1
+ end as htnwchf /* Hypertensive heart disease with heart failure */
+
+, case
+ when icd9_code = '40300' then 1
+ when icd9_code = '40310' then 1
+ when icd9_code = '40390' then 1
+ when icd9_code = '40501' then 1
+ when icd9_code = '40511' then 1
+ when icd9_code = '40591' then 1
+ when icd9_code between '64210' and '64214' then 1
+ end as hrenworf /* Hypertensive renal disease without renal failure */
+
+, case
+ when icd9_code = '40301' then 1
+ when icd9_code = '40311' then 1
+ when icd9_code = '40391' then 1
+ end as hrenwrf /* Hypertensive renal disease with renal failure */
+
+, case
+ when icd9_code = '40400' then 1
+ when icd9_code = '40410' then 1
+ when icd9_code = '40490' then 1
+ end as hhrwohrf /* Hypertensive heart and renal disease without heart or renal failure */
+
+, case
+ when icd9_code = '40401' then 1
+ when icd9_code = '40411' then 1
+ when icd9_code = '40491' then 1
+ end as hhrwchf /* Hypertensive heart and renal disease with heart failure */
+
+, case
+ when icd9_code = '40402' then 1
+ when icd9_code = '40412' then 1
+ when icd9_code = '40492' then 1
+ end as hhrwrf /* Hypertensive heart and renal disease with renal failure */
+
+, case
+ when icd9_code = '40403' then 1
+ when icd9_code = '40413' then 1
+ when icd9_code = '40493' then 1
+ end as hhrwhrf /* Hypertensive heart and renal disease with heart and renal failure */
+
+, case
+ when icd9_code between '64270' and '64274' then 1
+ when icd9_code between '64290' and '64294' then 1
+ end as ohtnpreg /* Other hypertension in pregnancy */
+
+ /******************** End Temporary Formats ***********************/
+
+, case
+ when icd9_code between '3420' and '3449' then 1
+ when icd9_code between '43820' and '43853' then 1
+ when icd9_code = '78072' then 1
+ end as para /* Paralysis */
+
+, case
+ when icd9_code between '3300' and '3319' then 1
+ when icd9_code = '3320' then 1
+ when icd9_code = '3334' then 1
+ when icd9_code = '3335' then 1
+ when icd9_code = '3337' then 1
+ when icd9_code in ('33371','33372','33379','33385','33394') then 1
+ when icd9_code between '3340' and '3359' then 1
+ when icd9_code = '3380' then 1
+ when icd9_code = '340' then 1
+ when icd9_code between '3411' and '3419' then 1
+ when icd9_code between '34500' and '34511' then 1
+ when icd9_code between '3452' and '3453' then 1
+ when icd9_code between '34540' and '34591' then 1
+ when icd9_code between '34700' and '34701' then 1
+ when icd9_code between '34710' and '34711' then 1
+ when icd9_code = '3483' then 1 -- discontinued icd-9
+ when icd9_code between '64940' and '64944' then 1
+ when icd9_code = '7687' then 1
+ when icd9_code between '76870' and '76873' then 1
+ when icd9_code = '7803' then 1
+ when icd9_code = '78031' then 1
+ when icd9_code = '78032' then 1
+ when icd9_code = '78033' then 1
+ when icd9_code = '78039' then 1
+ when icd9_code = '78097' then 1
+ when icd9_code = '7843' then 1
+ end as neuro /* Other neurological */
+
+, case
+ when icd9_code between '490' and '4928' then 1
+ when icd9_code between '49300' and '49392' then 1
+ when icd9_code between '494' and '4941' then 1
+ when icd9_code between '4950' and '505' then 1
+ when icd9_code = '5064' then 1
+ end as chrnlung /* Chronic pulmonary disease */
+
+, case
+ when icd9_code between '25000' and '25033' then 1
+ when icd9_code between '64800' and '64804' then 1
+ when icd9_code between '24900' and '24931' then 1
+ end as dm /* Diabetes w/o chronic complications*/
+
+, case
+ when icd9_code between '25040' and '25093' then 1
+ when icd9_code = '7751' then 1
+ when icd9_code between '24940' and '24991' then 1
+ end as dmcx /* Diabetes w/ chronic complications */
+
+, case
+ when icd9_code between '243' and '2442' then 1
+ when icd9_code = '2448' then 1
+ when icd9_code = '2449' then 1
+ end as hypothy /* Hypothyroidism */
+
+, case
+ when icd9_code = '585' then 1 -- discontinued code
+ when icd9_code = '5853' then 1
+ when icd9_code = '5854' then 1
+ when icd9_code = '5855' then 1
+ when icd9_code = '5856' then 1
+ when icd9_code = '5859' then 1
+ when icd9_code = '586' then 1
+ when icd9_code = 'V420' then 1
+ when icd9_code = 'V451' then 1
+ when icd9_code between 'V560' and 'V5632' then 1
+ when icd9_code = 'V568' then 1
+ when icd9_code between 'V4511' and 'V4512' then 1
+ end as renlfail /* Renal failure */
+
+, case
+ when icd9_code = '07022' then 1
+ when icd9_code = '07023' then 1
+ when icd9_code = '07032' then 1
+ when icd9_code = '07033' then 1
+ when icd9_code = '07044' then 1
+ when icd9_code = '07054' then 1
+ when icd9_code = '4560' then 1
+ when icd9_code = '4561' then 1
+ when icd9_code = '45620' then 1
+ when icd9_code = '45621' then 1
+ when icd9_code = '5710' then 1
+ when icd9_code = '5712' then 1
+ when icd9_code = '5713' then 1
+ when icd9_code between '57140' and '57149' then 1
+ when icd9_code = '5715' then 1
+ when icd9_code = '5716' then 1
+ when icd9_code = '5718' then 1
+ when icd9_code = '5719' then 1
+ when icd9_code = '5723' then 1
+ when icd9_code = '5728' then 1
+ when icd9_code = '5735' then 1
+ when icd9_code = 'V427' then 1
+ end as liver /* Liver disease */
+
+, case
+ when icd9_code = '53141' then 1
+ when icd9_code = '53151' then 1
+ when icd9_code = '53161' then 1
+ when icd9_code = '53170' then 1
+ when icd9_code = '53171' then 1
+ when icd9_code = '53191' then 1
+ when icd9_code = '53241' then 1
+ when icd9_code = '53251' then 1
+ when icd9_code = '53261' then 1
+ when icd9_code = '53270' then 1
+ when icd9_code = '53271' then 1
+ when icd9_code = '53291' then 1
+ when icd9_code = '53341' then 1
+ when icd9_code = '53351' then 1
+ when icd9_code = '53361' then 1
+ when icd9_code = '53370' then 1
+ when icd9_code = '53371' then 1
+ when icd9_code = '53391' then 1
+ when icd9_code = '53441' then 1
+ when icd9_code = '53451' then 1
+ when icd9_code = '53461' then 1
+ when icd9_code = '53470' then 1
+ when icd9_code = '53471' then 1
+ when icd9_code = '53491' then 1
+ end as ulcer /* Chronic Peptic ulcer disease (includes bleeding only if obstruction is also present) */
+
+, case
+ when icd9_code between '042' and '0449' then 1
+ end as aids /* HIV and AIDS */
+
+, case
+ when icd9_code between '20000' and '20238' then 1
+ when icd9_code between '20250' and '20301' then 1
+ when icd9_code = '2386' then 1
+ when icd9_code = '2733' then 1
+ when icd9_code between '20302' and '20382' then 1
+ end as lymph /* Lymphoma */
+
+, case
+ when icd9_code between '1960' and '1991' then 1
+ when icd9_code between '20970' and '20975' then 1
+ when icd9_code = '20979' then 1
+ when icd9_code = '78951' then 1
+ end as mets /* Metastatic cancer */
+
+, case
+ when icd9_code between '1400' and '1729' then 1
+ when icd9_code between '1740' and '1759' then 1
+ when icd9_code between '179' and '1958' then 1
+ when icd9_code between '20900' and '20924' then 1
+ when icd9_code between '20925' and '2093' then 1
+ when icd9_code between '20930' and '20936' then 1
+ when icd9_code between '25801' and '25803' then 1
+ end as tumor /* Solid tumor without metastasis */
+
+, case
+ when icd9_code = '7010' then 1
+ when icd9_code between '7100' and '7109' then 1
+ when icd9_code between '7140' and '7149' then 1
+ when icd9_code between '7200' and '7209' then 1
+ when icd9_code = '725' then 1
+ end as arth /* Rheumatoid arthritis/collagen vascular diseases */
+
+, case
+ when icd9_code between '2860' and '2869' then 1
+ when icd9_code = '2871' then 1
+ when icd9_code between '2873' and '2875' then 1
+ when icd9_code between '64930' and '64934' then 1
+ when icd9_code = '28984' then 1
+ end as coag /* Coagulation deficiency */
+
+, case
+ when icd9_code = '2780' then 1
+ when icd9_code = '27800' then 1
+ when icd9_code = '27801' then 1
+ when icd9_code = '27803' then 1
+ when icd9_code between '64910' and '64914' then 1
+ when icd9_code between 'V8530' and 'V8539' then 1
+ when icd9_code = 'V854' then 1 -- hierarchy used for AHRQ v3.6 and earlier
+ when icd9_code between 'V8541' and 'V8545' then 1
+ when icd9_code = 'V8554' then 1
+ when icd9_code = '79391' then 1
+ end as obese /* Obesity */
+
+, case
+ when icd9_code between '260' and '2639' then 1
+ when icd9_code between '78321' and '78322' then 1
+ end as wghtloss /* Weight loss */
+
+, case
+ when icd9_code between '2760' and '2769' then 1
+ end as lytes /* Fluid and electrolyte disorders - note:
+ this comorbidity should be dropped when
+ used with the AHRQ Patient Safety Indicators*/
+, case
+ when icd9_code = '2800' then 1
+ when icd9_code between '64820' and '64824' then 1
+ end as bldloss /* Blood loss anemia */
+
+, case
+ when icd9_code between '2801' and '2819' then 1
+ when icd9_code between '28521' and '28529' then 1
+ when icd9_code = '2859' then 1
+ end as anemdef /* Deficiency anemias */
+
+, case
+ when icd9_code between '2910' and '2913' then 1
+ when icd9_code = '2915' then 1
+ when icd9_code = '2918' then 1
+ when icd9_code = '29181' then 1
+ when icd9_code = '29182' then 1
+ when icd9_code = '29189' then 1
+ when icd9_code = '2919' then 1
+ when icd9_code between '30300' and '30393' then 1
+ when icd9_code between '30500' and '30503' then 1
+ end as alcohol /* Alcohol abuse */
+
+, case
+ when icd9_code = '2920' then 1
+ when icd9_code between '29282' and '29289' then 1
+ when icd9_code = '2929' then 1
+ when icd9_code between '30400' and '30493' then 1
+ when icd9_code between '30520' and '30593' then 1
+ when icd9_code between '64830' and '64834' then 1
+ end as drug /* Drug abuse */
+
+, case
+ when icd9_code between '29500' and '2989' then 1
+ when icd9_code = '29910' then 1
+ when icd9_code = '29911' then 1
+ end as psych /* Psychoses */
+
+, case
+ when icd9_code = '3004' then 1
+ when icd9_code = '30112' then 1
+ when icd9_code = '3090' then 1
+ when icd9_code = '3091' then 1
+ when icd9_code = '311' then 1
+ end as depress /* Depression */
+from diagnoses_icd icd
+where seq_num = 1
+)
+-- collapse the icd9_code specific flags into hadm_id specific flags
+-- this groups comorbidities together for a single patient admission
+, eligrp as
+(
+ select hadm_id
+ , max(chf) as chf
+ , max(arythm) as arythm
+ , max(valve) as valve
+ , max(pulmcirc) as pulmcirc
+ , max(perivasc) as perivasc
+ , max(htn) as htn
+ , max(htncx) as htncx
+ , max(htnpreg) as htnpreg
+ , max(htnwochf) as htnwochf
+ , max(htnwchf) as htnwchf
+ , max(hrenworf) as hrenworf
+ , max(hrenwrf) as hrenwrf
+ , max(hhrwohrf) as hhrwohrf
+ , max(hhrwchf) as hhrwchf
+ , max(hhrwrf) as hhrwrf
+ , max(hhrwhrf) as hhrwhrf
+ , max(ohtnpreg) as ohtnpreg
+ , max(para) as para
+ , max(neuro) as neuro
+ , max(chrnlung) as chrnlung
+ , max(dm) as dm
+ , max(dmcx) as dmcx
+ , max(hypothy) as hypothy
+ , max(renlfail) as renlfail
+ , max(liver) as liver
+ , max(ulcer) as ulcer
+ , max(aids) as aids
+ , max(lymph) as lymph
+ , max(mets) as mets
+ , max(tumor) as tumor
+ , max(arth) as arth
+ , max(coag) as coag
+ , max(obese) as obese
+ , max(wghtloss) as wghtloss
+ , max(lytes) as lytes
+ , max(bldloss) as bldloss
+ , max(anemdef) as anemdef
+ , max(alcohol) as alcohol
+ , max(drug) as drug
+ , max(psych) as psych
+ , max(depress) as depress
+from eliflg
+group by hadm_id
+)
+-- now merge these flags together to define elixhauser
+-- most are straightforward.. but hypertension flags are a bit more complicated
+select adm.subject_id, adm.hadm_id
+, case
+ when chf = 1 then 1
+ when htnwchf = 1 then 1
+ when hhrwchf = 1 then 1
+ when hhrwhrf = 1 then 1
+ else 0 end as congestive_heart_failure
+, case
+ when arythm = 1 then 1
+ else 0 end as cardiac_arrhythmias
+, case when valve = 1 then 1 else 0 end as valvular_disease
+, case when pulmcirc = 1 then 1 else 0 end as pulmonary_circulation
+, case when perivasc = 1 then 1 else 0 end as peripheral_vascular
+
+-- we combine "htn" and "htncx" into "HYPERTENSION"
+-- note "htn" (hypertension) is only 1 if "htncx" (complicated hypertension) is 0
+-- this matters if you filter on DRG but for this query we can just merge them immediately
+, case
+ when htn = 1 then 1
+ when htncx = 1 then 1
+ when htnpreg = 1 then 1
+ when htnwochf = 1 then 1
+ when htnwchf = 1 then 1
+ when hrenworf = 1 then 1
+ when hrenwrf = 1 then 1
+ when hhrwohrf = 1 then 1
+ when hhrwchf = 1 then 1
+ when hhrwrf = 1 then 1
+ when hhrwhrf = 1 then 1
+ when ohtnpreg = 1 then 1
+ else 0 end as hypertension
+
+, case when para = 1 then 1 else 0 end as paralysis
+, case when neuro = 1 then 1 else 0 end as other_neurological
+, case when chrnlung = 1 then 1 else 0 end as chronic_pulmonary
+, case
+ -- only the more severe comorbidity (complicated diabetes) is kept
+ when dmcx = 1 then 0
+ when dm = 1 then 1
+ else 0 end as diabetes_uncomplicated
+, case when dmcx = 1 then 1 else 0 end as diabetes_complicated
+, case when hypothy = 1 then 1 else 0 end as hypothyroidism
+, case
+ when renlfail = 1 then 1
+ when hrenwrf = 1 then 1
+ when hhrwrf = 1 then 1
+ when hhrwhrf = 1 then 1
+ else 0 end as renal_failure
+
+, case when liver = 1 then 1 else 0 end as liver_disease
+, case when ulcer = 1 then 1 else 0 end as peptic_ulcer
+, case when aids = 1 then 1 else 0 end as aids
+, case when lymph = 1 then 1 else 0 end as lymphoma
+, case when mets = 1 then 1 else 0 end as metastatic_cancer
+, case
+ -- only the more severe comorbidity (metastatic cancer) is kept
+ when mets = 1 then 0
+ when tumor = 1 then 1
+ else 0 end as solid_tumor
+, case when arth = 1 then 1 else 0 end as rheumatoid_arthritis
+, case when coag = 1 then 1 else 0 end as coagulopathy
+, case when obese = 1 then 1 else 0 end as obesity
+, case when wghtloss = 1 then 1 else 0 end as weight_loss
+, case when lytes = 1 then 1 else 0 end as fluid_electrolyte
+, case when bldloss = 1 then 1 else 0 end as blood_loss_anemia
+, case when anemdef = 1 then 1 else 0 end as deficiency_anemias
+, case when alcohol = 1 then 1 else 0 end as alcohol_abuse
+, case when drug = 1 then 1 else 0 end as drug_abuse
+, case when psych = 1 then 1 else 0 end as psychoses
+, case when depress = 1 then 1 else 0 end as depression
+
+from admissions adm
+left join eligrp eli
+ on adm.hadm_id = eli.hadm_id
+order by adm.hadm_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/17_elixhauser_quan.sql b/mimic-iii-carevue/code/concepts/17_elixhauser_quan.sql
new file mode 100644
index 000000000..1f67db01c
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/17_elixhauser_quan.sql
@@ -0,0 +1,271 @@
+drop table if exists elixhauser_quan; create table elixhauser_quan as
+-- This code calculates the Elixhauser comorbidities as defined in Quan et. al 2009:
+-- Quan, Hude, et al. "Coding algorithms for defining comorbidities in
+-- ICD-9-CM and ICD-10 administrative data." Medical care (2005): 1130-1139.
+-- https://www.ncbi.nlm.nih.gov/pubmed/16224307
+
+-- Quan defined an "Enhanced ICD-9" coding scheme for deriving Elixhauser
+-- comorbidities from ICD-9 billing codes. This script implements that calculation.
+
+-- The logic of the code is roughly that, if the comorbidity lists a length 3
+-- ICD-9 code (e.g. 585), then we only require a match on the first 3 characters.
+
+-- This code derives each comorbidity as follows:
+-- 1) ICD9_CODE is directly compared to 5 character codes
+-- 2) The first 4 characters of ICD9_CODE are compared to 4 character codes
+-- 3) The first 3 characters of ICD9_CODE are compared to 3 character codes
+with eliflg as
+(
+select hadm_id, seq_num, icd9_code
+, case
+ when icd9_code in ('39891','40201','40211','40291','40401','40403','40411','40413','40491','40493') then 1
+ when substr(icd9_code, 1, 4) in ('4254','4255','4257','4258','4259') then 1
+ when substr(icd9_code, 1, 3) in ('428') then 1
+ else 0 end as chf /* Congestive heart failure */
+
+, case
+ when icd9_code in ('42613','42610','42612','99601','99604') then 1
+ when substr(icd9_code, 1, 4) in ('4260','4267','4269','4270','4271','4272','4273','4274','4276','4278','4279','7850','V450','V533') then 1
+ else 0 end as arrhy
+
+, case
+ when substr(icd9_code, 1, 4) in ('0932','7463','7464','7465','7466','V422','V433') then 1
+ when substr(icd9_code, 1, 3) in ('394','395','396','397','424') then 1
+ else 0 end as valve /* Valvular disease */
+
+, case
+ when substr(icd9_code, 1, 4) in ('4150','4151','4170','4178','4179') then 1
+ when substr(icd9_code, 1, 3) in ('416') then 1
+ else 0 end as pulmcirc /* Pulmonary circulation disorder */
+
+, case
+ when substr(icd9_code, 1, 4) in ('0930','4373','4431','4432','4438','4439','4471','5571','5579','V434') then 1
+ when substr(icd9_code, 1, 3) in ('440','441') then 1
+ else 0 end as perivasc /* Peripheral vascular disorder */
+
+, case
+ when substr(icd9_code, 1, 3) in ('401') then 1
+ else 0 end as htn /* Hypertension, uncomplicated */
+
+, case
+ when substr(icd9_code, 1, 3) in ('402','403','404','405') then 1
+ else 0 end as htncx /* Hypertension, complicated */
+
+, case
+ when substr(icd9_code, 1, 4) in ('3341','3440','3441','3442','3443','3444','3445','3446','3449') then 1
+ when substr(icd9_code, 1, 3) in ('342','343') then 1
+ else 0 end as para /* Paralysis */
+
+, case
+ when icd9_code in ('33392') then 1
+ when substr(icd9_code, 1, 4) in ('3319','3320','3321','3334','3335','3362','3481','3483','7803','7843') then 1
+ when substr(icd9_code, 1, 3) in ('334','335','340','341','345') then 1
+ else 0 end as neuro /* Other neurological */
+
+, case
+ when substr(icd9_code, 1, 4) in ('4168','4169','5064','5081','5088') then 1
+ when substr(icd9_code, 1, 3) in ('490','491','492','493','494','495','496','500','501','502','503','504','505') then 1
+ else 0 end as chrnlung /* Chronic pulmonary disease */
+
+, case
+ when substr(icd9_code, 1, 4) in ('2500','2501','2502','2503') then 1
+ else 0 end as dm /* Diabetes w/o chronic complications*/
+
+, case
+ when substr(icd9_code, 1, 4) in ('2504','2505','2506','2507','2508','2509') then 1
+ else 0 end as dmcx /* Diabetes w/ chronic complications */
+
+, case
+ when substr(icd9_code, 1, 4) in ('2409','2461','2468') then 1
+ when substr(icd9_code, 1, 3) in ('243','244') then 1
+ else 0 end as hypothy /* Hypothyroidism */
+
+, case
+ when icd9_code in ('40301','40311','40391','40402','40403','40412','40413','40492','40493') then 1
+ when substr(icd9_code, 1, 4) in ('5880','V420','V451') then 1
+ when substr(icd9_code, 1, 3) in ('585','586','V56') then 1
+ else 0 end as renlfail /* Renal failure */
+
+, case
+ when icd9_code in ('07022','07023','07032','07033','07044','07054') then 1
+ when substr(icd9_code, 1, 4) in ('0706','0709','4560','4561','4562','5722','5723','5724','5728','5733','5734','5738','5739','V427') then 1
+ when substr(icd9_code, 1, 3) in ('570','571') then 1
+ else 0 end as liver /* Liver disease */
+
+, case
+ when substr(icd9_code, 1, 4) in ('5317','5319','5327','5329','5337','5339','5347','5349') then 1
+ else 0 end as ulcer /* Chronic Peptic ulcer disease (includes bleeding only if obstruction is also present) */
+
+, case
+ when substr(icd9_code, 1, 3) in ('042','043','044') then 1
+ else 0 end as aids /* HIV and AIDS */
+
+, case
+ when substr(icd9_code, 1, 4) in ('2030','2386') then 1
+ when substr(icd9_code, 1, 3) in ('200','201','202') then 1
+ else 0 end as lymph /* Lymphoma */
+
+, case
+ when substr(icd9_code, 1, 3) in ('196','197','198','199') then 1
+ else 0 end as mets /* Metastatic cancer */
+
+, case
+ when substr(icd9_code, 1, 3) in
+ (
+ '140','141','142','143','144','145','146','147','148','149','150','151','152'
+ ,'153','154','155','156','157','158','159','160','161','162','163','164','165'
+ ,'166','167','168','169','170','171','172','174','175','176','177','178','179'
+ ,'180','181','182','183','184','185','186','187','188','189','190','191','192'
+ ,'193','194','195'
+ ) then 1
+ else 0 end as tumor /* Solid tumor without metastasis */
+
+, case
+ when icd9_code in ('72889','72930') then 1
+ when substr(icd9_code, 1, 4) in ('7010','7100','7101','7102','7103','7104','7108','7109','7112','7193','7285') then 1
+ when substr(icd9_code, 1, 3) in ('446','714','720','725') then 1
+ else 0 end as arth /* Rheumatoid arthritis/collagen vascular diseases */
+
+, case
+ when substr(icd9_code, 1, 4) in ('2871','2873','2874','2875') then 1
+ when substr(icd9_code, 1, 3) in ('286') then 1
+ else 0 end as coag /* Coagulation deficiency */
+
+, case
+ when substr(icd9_code, 1, 4) in ('2780') then 1
+ else 0 end as obese /* Obesity */
+
+, case
+ when substr(icd9_code, 1, 4) in ('7832','7994') then 1
+ when substr(icd9_code, 1, 3) in ('260','261','262','263') then 1
+ else 0 end as wghtloss /* Weight loss */
+
+, case
+ when substr(icd9_code, 1, 4) in ('2536') then 1
+ when substr(icd9_code, 1, 3) in ('276') then 1
+ else 0 end as lytes /* Fluid and electrolyte disorders */
+
+, case
+ when substr(icd9_code, 1, 4) in ('2800') then 1
+ else 0 end as bldloss /* Blood loss anemia */
+
+, case
+ when substr(icd9_code, 1, 4) in ('2801','2808','2809') then 1
+ when substr(icd9_code, 1, 3) in ('281') then 1
+ else 0 end as anemdef /* Deficiency anemias */
+
+, case
+ when substr(icd9_code, 1, 4) in ('2652','2911','2912','2913','2915','2918','2919','3030','3039','3050','3575','4255','5353','5710','5711','5712','5713','V113') then 1
+ when substr(icd9_code, 1, 3) in ('980') then 1
+ else 0 end as alcohol /* Alcohol abuse */
+
+, case
+ when icd9_code in ('V6542') then 1
+ when substr(icd9_code, 1, 4) in ('3052','3053','3054','3055','3056','3057','3058','3059') then 1
+ when substr(icd9_code, 1, 3) in ('292','304') then 1
+ else 0 end as drug /* Drug abuse */
+
+, case
+ when icd9_code in ('29604','29614','29644','29654') then 1
+ when substr(icd9_code, 1, 4) in ('2938') then 1
+ when substr(icd9_code, 1, 3) in ('295','297','298') then 1
+ else 0 end as psych /* Psychoses */
+
+, case
+ when substr(icd9_code, 1, 4) in ('2962','2963','2965','3004') then 1
+ when substr(icd9_code, 1, 3) in ('309','311') then 1
+ else 0 end as depress /* Depression */
+from diagnoses_icd icd
+where seq_num != 1 -- we do not include the primary icd-9 code
+)
+-- collapse the icd9_code specific flags into hadm_id specific flags
+-- this groups comorbidities together for a single patient admission
+, eligrp as
+(
+ select hadm_id
+ , max(chf) as chf
+ , max(arrhy) as arrhy
+ , max(valve) as valve
+ , max(pulmcirc) as pulmcirc
+ , max(perivasc) as perivasc
+ , max(htn) as htn
+ , max(htncx) as htncx
+ , max(para) as para
+ , max(neuro) as neuro
+ , max(chrnlung) as chrnlung
+ , max(dm) as dm
+ , max(dmcx) as dmcx
+ , max(hypothy) as hypothy
+ , max(renlfail) as renlfail
+ , max(liver) as liver
+ , max(ulcer) as ulcer
+ , max(aids) as aids
+ , max(lymph) as lymph
+ , max(mets) as mets
+ , max(tumor) as tumor
+ , max(arth) as arth
+ , max(coag) as coag
+ , max(obese) as obese
+ , max(wghtloss) as wghtloss
+ , max(lytes) as lytes
+ , max(bldloss) as bldloss
+ , max(anemdef) as anemdef
+ , max(alcohol) as alcohol
+ , max(drug) as drug
+ , max(psych) as psych
+ , max(depress) as depress
+from eliflg
+group by hadm_id
+)
+-- now merge these flags together to define elixhauser
+-- most are straightforward.. but hypertension flags are a bit more complicated
+
+
+select adm.hadm_id
+, chf as congestive_heart_failure
+, arrhy as cardiac_arrhythmias
+, valve as valvular_disease
+, pulmcirc as pulmonary_circulation
+, perivasc as peripheral_vascular
+-- we combine "htn" and "htncx" into "HYPERTENSION"
+, case
+ when htn = 1 then 1
+ when htncx = 1 then 1
+ else 0 end as hypertension
+, para as paralysis
+, neuro as other_neurological
+, chrnlung as chronic_pulmonary
+-- only the more severe comorbidity (complicated diabetes) is kept
+, case
+ when dmcx = 1 then 0
+ when dm = 1 then 1
+ else 0 end as diabetes_uncomplicated
+, dmcx as diabetes_complicated
+, hypothy as hypothyroidism
+, renlfail as renal_failure
+, liver as liver_disease
+, ulcer as peptic_ulcer
+, aids as aids
+, lymph as lymphoma
+, mets as metastatic_cancer
+-- only the more severe comorbidity (metastatic cancer) is kept
+, case
+ when mets = 1 then 0
+ when tumor = 1 then 1
+ else 0 end as solid_tumor
+, arth as rheumatoid_arthritis
+, coag as coagulopathy
+, obese as obesity
+, wghtloss as weight_loss
+, lytes as fluid_electrolyte
+, bldloss as blood_loss_anemia
+, anemdef as deficiency_anemias
+, alcohol as alcohol_abuse
+, drug as drug_abuse
+, psych as psychoses
+, depress as depression
+
+from admissions adm
+left join eligrp eli
+ on adm.hadm_id = eli.hadm_id
+order by adm.hadm_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/18_elixhauser_score_ahrq.sql b/mimic-iii-carevue/code/concepts/18_elixhauser_score_ahrq.sql
new file mode 100644
index 000000000..4258cbc17
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/18_elixhauser_score_ahrq.sql
@@ -0,0 +1,107 @@
+drop table if exists elixhauser_score_ahrq; create table elixhauser_score_ahrq as
+-- This query provides various methods of combining the Elixhauser components into a single score
+-- The methods are called "vanWalRaven" and "SID30", and "SID29"
+select subject_id, hadm_id
+, -- Below is the van Walraven score
+ 0 * aids +
+ 0 * alcohol_abuse +
+ -2 * blood_loss_anemia +
+ 7 * congestive_heart_failure +
+ -- Cardiac arrhythmias are not included in van Walraven based on Quan 2007
+ 3 * chronic_pulmonary +
+ 3 * coagulopathy +
+ -2 * deficiency_anemias +
+ -3 * depression +
+ 0 * diabetes_complicated +
+ 0 * diabetes_uncomplicated +
+ -7 * drug_abuse +
+ 5 * fluid_electrolyte +
+ 0 * hypertension +
+ 0 * hypothyroidism +
+ 11 * liver_disease +
+ 9 * lymphoma +
+ 12 * metastatic_cancer +
+ 6 * other_neurological +
+ -4 * obesity +
+ 7 * paralysis +
+ 2 * peripheral_vascular +
+ 0 * peptic_ulcer +
+ 0 * psychoses +
+ 4 * pulmonary_circulation +
+ 0 * rheumatoid_arthritis +
+ 5 * renal_failure +
+ 4 * solid_tumor +
+ -1 * valvular_disease +
+ 6 * weight_loss
+as elixhauser_vanwalraven
+
+
+
+, -- Below is the 29 component SID score
+ 0 * aids +
+ -2 * alcohol_abuse +
+ -2 * blood_loss_anemia +
+ -- Cardiac arrhythmias are not included in SID-29
+ 9 * congestive_heart_failure +
+ 3 * chronic_pulmonary +
+ 9 * coagulopathy +
+ 0 * deficiency_anemias +
+ -4 * depression +
+ 0 * diabetes_complicated +
+ -1 * diabetes_uncomplicated +
+ -8 * drug_abuse +
+ 9 * fluid_electrolyte +
+ -1 * hypertension +
+ 0 * hypothyroidism +
+ 5 * liver_disease +
+ 6 * lymphoma +
+ 13 * metastatic_cancer +
+ 4 * other_neurological +
+ -4 * obesity +
+ 3 * paralysis +
+ 0 * peptic_ulcer +
+ 4 * peripheral_vascular +
+ -4 * psychoses +
+ 5 * pulmonary_circulation +
+ 6 * renal_failure +
+ 0 * rheumatoid_arthritis +
+ 8 * solid_tumor +
+ 0 * valvular_disease +
+ 8 * weight_loss
+as elixhauser_SID29
+
+
+, -- Below is the 30 component SID score
+ 0 * aids +
+ 0 * alcohol_abuse +
+ -3 * blood_loss_anemia +
+ 8 * cardiac_arrhythmias +
+ 9 * congestive_heart_failure +
+ 3 * chronic_pulmonary +
+ 12 * coagulopathy +
+ 0 * deficiency_anemias +
+ -5 * depression +
+ 1 * diabetes_complicated +
+ 0 * diabetes_uncomplicated +
+ -11 * drug_abuse +
+ 11 * fluid_electrolyte +
+ -2 * hypertension +
+ 0 * hypothyroidism +
+ 7 * liver_disease +
+ 8 * lymphoma +
+ 17 * metastatic_cancer +
+ 5 * other_neurological +
+ -5 * obesity +
+ 4 * paralysis +
+ 0 * peptic_ulcer +
+ 4 * peripheral_vascular +
+ -6 * psychoses +
+ 5 * pulmonary_circulation +
+ 7 * renal_failure +
+ 0 * rheumatoid_arthritis +
+ 10 * solid_tumor +
+ 0 * valvular_disease +
+ 10 * weight_loss
+as elixhauser_sid30
+
+from elixhauser_ahrq_v37;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/19_elixhauser_score_quan.sql b/mimic-iii-carevue/code/concepts/19_elixhauser_score_quan.sql
new file mode 100644
index 000000000..b0d506284
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/19_elixhauser_score_quan.sql
@@ -0,0 +1,108 @@
+drop table if exists elixhauser_score_quan; create table elixhauser_score_quan as
+-- This query provides various methods of combining the Elixhauser components into a single score
+-- The methods are called "vanWalRaven" and "SID30", and "SID29"
+
+select hadm_id
+, -- Below is the van Walraven score
+ 0 * aids +
+ 0 * alcohol_abuse +
+ -2 * blood_loss_anemia +
+ 7 * congestive_heart_failure +
+ -- Cardiac arrhythmias are not included in van Walraven based on Quan 2007
+ 3 * chronic_pulmonary +
+ 3 * coagulopathy +
+ -2 * deficiency_anemias +
+ -3 * depression +
+ 0 * diabetes_complicated +
+ 0 * diabetes_uncomplicated +
+ -7 * drug_abuse +
+ 5 * fluid_electrolyte +
+ 0 * hypertension +
+ 0 * hypothyroidism +
+ 11 * liver_disease +
+ 9 * lymphoma +
+ 12 * metastatic_cancer +
+ 6 * other_neurological +
+ -4 * obesity +
+ 7 * paralysis +
+ 2 * peripheral_vascular +
+ 0 * peptic_ulcer +
+ 0 * psychoses +
+ 4 * pulmonary_circulation +
+ 0 * rheumatoid_arthritis +
+ 5 * renal_failure +
+ 4 * solid_tumor +
+ -1 * valvular_disease +
+ 6 * weight_loss
+as elixhauser_vanwalraven
+
+
+
+, -- Below is the 29 component SID score
+ 0 * aids +
+ -2 * alcohol_abuse +
+ -2 * blood_loss_anemia +
+ -- Cardiac arrhythmias are not included in SID-29
+ 9 * congestive_heart_failure +
+ 3 * chronic_pulmonary +
+ 9 * coagulopathy +
+ 0 * deficiency_anemias +
+ -4 * depression +
+ 0 * diabetes_complicated +
+ -1 * diabetes_uncomplicated +
+ -8 * drug_abuse +
+ 9 * fluid_electrolyte +
+ -1 * hypertension +
+ 0 * hypothyroidism +
+ 5 * liver_disease +
+ 6 * lymphoma +
+ 13 * metastatic_cancer +
+ 4 * other_neurological +
+ -4 * obesity +
+ 3 * paralysis +
+ 0 * peptic_ulcer +
+ 4 * peripheral_vascular +
+ -4 * psychoses +
+ 5 * pulmonary_circulation +
+ 6 * renal_failure +
+ 0 * rheumatoid_arthritis +
+ 8 * solid_tumor +
+ 0 * valvular_disease +
+ 8 * weight_loss
+as elixhauser_SID29
+
+
+, -- Below is the 30 component SID score
+ 0 * aids +
+ 0 * alcohol_abuse +
+ -3 * blood_loss_anemia +
+ 8 * cardiac_arrhythmias +
+ 9 * congestive_heart_failure +
+ 3 * chronic_pulmonary +
+ 12 * coagulopathy +
+ 0 * deficiency_anemias +
+ -5 * depression +
+ 1 * diabetes_complicated +
+ 0 * diabetes_uncomplicated +
+ -11 * drug_abuse +
+ 11 * fluid_electrolyte +
+ -2 * hypertension +
+ 0 * hypothyroidism +
+ 7 * liver_disease +
+ 8 * lymphoma +
+ 17 * metastatic_cancer +
+ 5 * other_neurological +
+ -5 * obesity +
+ 4 * paralysis +
+ 0 * peptic_ulcer +
+ 4 * peripheral_vascular +
+ -6 * psychoses +
+ 5 * pulmonary_circulation +
+ 7 * renal_failure +
+ 0 * rheumatoid_arthritis +
+ 10 * solid_tumor +
+ 0 * valvular_disease +
+ 10 * weight_loss
+as elixhauser_sid30
+
+from elixhauser_quan;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/20_icustay_detail.sql b/mimic-iii-carevue/code/concepts/20_icustay_detail.sql
new file mode 100644
index 000000000..6c679dc00
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/20_icustay_detail.sql
@@ -0,0 +1,97 @@
+drop table if exists icustay_detail; create table icustay_detail as
+
+-- This query extracts useful demographic/administrative information for patient ICU stays
+
+select ie.subject_id, ie.hadm_id, ie.icustay_id
+
+-- patient level factors
+, pat.gender, pat.dod
+
+-- hospital level factors
+, adm.admittime, adm.dischtime
+, round((cast(extract(epoch from (adm.dischtime - adm.admittime))/(60*60*24) as numeric)), 8) as los_hospital
+, round((cast(extract(epoch from (ie.intime - pat.dob))/(60*60*24*365) as numeric)), 8) as admission_age
+, adm.ethnicity
+, case when ethnicity in
+ (
+ 'WHITE' -- 40996
+ , 'WHITE - RUSSIAN' -- 164
+ , 'WHITE - OTHER EUROPEAN' -- 81
+ , 'WHITE - BRAZILIAN' -- 59
+ , 'WHITE - EASTERN EUROPEAN' -- 25
+ ) then 'white'
+ when ethnicity in
+ (
+ 'BLACK/AFRICAN AMERICAN' -- 5440
+ , 'BLACK/CAPE VERDEAN' -- 200
+ , 'BLACK/HAITIAN' -- 101
+ , 'BLACK/AFRICAN' -- 44
+ , 'CARIBBEAN ISLAND' -- 9
+ ) then 'black'
+ when ethnicity in
+ (
+ 'HISPANIC OR LATINO' -- 1696
+ , 'HISPANIC/LATINO - PUERTO RICAN' -- 232
+ , 'HISPANIC/LATINO - DOMINICAN' -- 78
+ , 'HISPANIC/LATINO - GUATEMALAN' -- 40
+ , 'HISPANIC/LATINO - CUBAN' -- 24
+ , 'HISPANIC/LATINO - SALVADORAN' -- 19
+ , 'HISPANIC/LATINO - CENTRAL AMERICAN (OTHER)' -- 13
+ , 'HISPANIC/LATINO - MEXICAN' -- 13
+ , 'HISPANIC/LATINO - COLOMBIAN' -- 9
+ , 'HISPANIC/LATINO - HONDURAN' -- 4
+ ) then 'hispanic'
+ when ethnicity in
+ (
+ 'ASIAN' -- 1509
+ , 'ASIAN - CHINESE' -- 277
+ , 'ASIAN - ASIAN INDIAN' -- 85
+ , 'ASIAN - VIETNAMESE' -- 53
+ , 'ASIAN - FILIPINO' -- 25
+ , 'ASIAN - CAMBODIAN' -- 17
+ , 'ASIAN - OTHER' -- 17
+ , 'ASIAN - KOREAN' -- 13
+ , 'ASIAN - JAPANESE' -- 7
+ , 'ASIAN - THAI' -- 4
+ ) then 'asian'
+ when ethnicity in
+ (
+ 'AMERICAN INDIAN/ALASKA NATIVE' -- 51
+ , 'AMERICAN INDIAN/ALASKA NATIVE FEDERALLY RECOGNIZED TRIBE' -- 3
+ ) then 'native'
+ when ethnicity in
+ (
+ 'UNKNOWN/NOT SPECIFIED' -- 4523
+ , 'UNABLE TO OBTAIN' -- 814
+ , 'PATIENT DECLINED TO ANSWER' -- 559
+ ) then 'unknown'
+ else 'other' end as ethnicity_grouped
+ -- , 'OTHER' -- 1512
+ -- , 'MULTI RACE ETHNICITY' -- 130
+ -- , 'PORTUGUESE' -- 61
+ -- , 'MIDDLE EASTERN' -- 43
+ -- , 'NATIVE HAWAIIAN OR OTHER PACIFIC ISLANDER' -- 18
+ -- , 'SOUTH AMERICAN' -- 8
+, adm.hospital_expire_flag
+, dense_rank() over (partition by adm.subject_id order by adm.admittime) as hospstay_seq
+, case
+ when dense_rank() over (partition by adm.subject_id order by adm.admittime) = 1 then true
+ else false end as first_hosp_stay
+
+-- icu level factors
+, ie.intime, ie.outtime
+, round((cast(extract(epoch from (ie.outtime - ie.intime))/(60*60*24) as numeric)), 8) as los_icu
+, dense_rank() over (partition by ie.hadm_id order by ie.intime) as icustay_seq
+
+-- first ICU stay *for the current hospitalization*
+, case
+ when dense_rank() over (partition by ie.hadm_id order by ie.intime) = 1 then true
+ else false end as first_icu_stay
+
+from icustays ie
+inner join admissions adm
+ on ie.hadm_id = adm.hadm_id
+inner join patients pat
+ on ie.subject_id = pat.subject_id
+where adm.has_chartevents_data = 1
+order by ie.subject_id, adm.admittime, ie.intime;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/21_blood_gas_first_day.sql b/mimic-iii-carevue/code/concepts/21_blood_gas_first_day.sql
new file mode 100644
index 000000000..d08299601
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/21_blood_gas_first_day.sql
@@ -0,0 +1,106 @@
+drop table if exists blood_gas_first_day; create table blood_gas_first_day as
+-- The aim of this query is to pivot entries related to blood gases and
+-- chemistry values which were found in labevents
+
+-- things to check:
+-- when a mixed venous/arterial blood sample are taken at the same time, is the store time different?
+
+with pvt as
+( -- begin query that extracts the data
+ select ie.subject_id, ie.hadm_id, ie.icustay_id
+ -- here we assign labels to itemids
+ -- this also fuses together multiple itemids containing the same data
+ , case
+ when itemid = 50800 then 'SPECIMEN'
+ when itemid = 50801 then 'AADO2'
+ when itemid = 50802 then 'BASEEXCESS'
+ when itemid = 50803 then 'BICARBONATE'
+ when itemid = 50804 then 'TOTALCO2'
+ when itemid = 50805 then 'CARBOXYHEMOGLOBIN'
+ when itemid = 50806 then 'CHLORIDE'
+ when itemid = 50808 then 'CALCIUM'
+ when itemid = 50809 then 'GLUCOSE'
+ when itemid = 50810 then 'HEMATOCRIT'
+ when itemid = 50811 then 'HEMOGLOBIN'
+ when itemid = 50812 then 'INTUBATED'
+ when itemid = 50813 then 'LACTATE'
+ when itemid = 50814 then 'METHEMOGLOBIN'
+ when itemid = 50815 then 'O2FLOW'
+ when itemid = 50816 then 'FIO2'
+ when itemid = 50817 then 'SO2' -- oxygensaturation
+ when itemid = 50818 then 'PCO2'
+ when itemid = 50819 then 'PEEP'
+ when itemid = 50820 then 'PH'
+ when itemid = 50821 then 'PO2'
+ when itemid = 50822 then 'POTASSIUM'
+ when itemid = 50823 then 'REQUIREDO2'
+ when itemid = 50824 then 'SODIUM'
+ when itemid = 50825 then 'TEMPERATURE'
+ when itemid = 50826 then 'TIDALVOLUME'
+ when itemid = 50827 then 'VENTILATIONRATE'
+ when itemid = 50828 then 'VENTILATOR'
+ else null
+ end as label
+ , charttime
+ , value
+ -- add in some sanity checks on the values
+ , case
+ when valuenum <= 0 and itemid != 50802 then null -- allow negative baseexcess
+ when itemid = 50810 and valuenum > 100 then null -- hematocrit
+ -- ensure FiO2 is a valid number between 21-100
+ -- mistakes are rare (<100 obs out of ~100,000)
+ -- there are 862 obs of valuenum == 20 - some people round down!
+ -- rather than risk imputing garbage data for FiO2, we simply null invalid values
+ when itemid = 50816 and valuenum < 20 then null
+ when itemid = 50816 and valuenum > 100 then null
+ when itemid = 50817 and valuenum > 100 then null -- O2 sat
+ when itemid = 50815 and valuenum > 70 then null -- O2 flow
+ when itemid = 50821 and valuenum > 800 then null -- PO2
+ -- conservative upper limit
+ else valuenum
+ end as valuenum
+ from icustays ie
+ left join labevents le
+ on le.subject_id = ie.subject_id and le.hadm_id = ie.hadm_id
+ and le.charttime between (ie.intime - interval '6 hour') and (ie.intime + interval '1 day')
+ and le.itemid in
+ -- blood gases
+ (
+ 50800, 50801, 50802, 50803, 50804, 50805, 50806, 50807, 50808, 50809
+ , 50810, 50811, 50812, 50813, 50814, 50815, 50816, 50817, 50818, 50819
+ , 50820, 50821, 50822, 50823, 50824, 50825, 50826, 50827, 50828
+ , 51545
+ )
+)
+select pvt.subject_id, pvt.hadm_id, pvt.icustay_id, pvt.charttime
+, max(case when label = 'SPECIMEN' then value else null end) as specimen
+, max(case when label = 'AADO2' then valuenum else null end) as aado2
+, max(case when label = 'BASEEXCESS' then valuenum else null end) as baseexcess
+, max(case when label = 'BICARBONATE' then valuenum else null end) as bicarbonate
+, max(case when label = 'TOTALCO2' then valuenum else null end) as totalco2
+, max(case when label = 'CARBOXYHEMOGLOBIN' then valuenum else null end) as carboxyhemoglobin
+, max(case when label = 'CHLORIDE' then valuenum else null end) as chloride
+, max(case when label = 'CALCIUM' then valuenum else null end) as calcium
+, max(case when label = 'GLUCOSE' then valuenum else null end) as glucose
+, max(case when label = 'HEMATOCRIT' then valuenum else null end) as hematocrit
+, max(case when label = 'HEMOGLOBIN' then valuenum else null end) as hemoglobin
+, max(case when label = 'INTUBATED' then valuenum else null end) as intubated
+, max(case when label = 'LACTATE' then valuenum else null end) as lactate
+, max(case when label = 'METHEMOGLOBIN' then valuenum else null end) as methemoglobin
+, max(case when label = 'O2FLOW' then valuenum else null end) as o2flow
+, max(case when label = 'FIO2' then valuenum else null end) as fio2
+, max(case when label = 'SO2' then valuenum else null end) as so2 -- oxygensaturation
+, max(case when label = 'PCO2' then valuenum else null end) as pco2
+, max(case when label = 'PEEP' then valuenum else null end) as peep
+, max(case when label = 'PH' then valuenum else null end) as ph
+, max(case when label = 'PO2' then valuenum else null end) as po2
+, max(case when label = 'POTASSIUM' then valuenum else null end) as potassium
+, max(case when label = 'REQUIREDO2' then valuenum else null end) as requiredo2
+, max(case when label = 'SODIUM' then valuenum else null end) as sodium
+, max(case when label = 'TEMPERATURE' then valuenum else null end) as temperature
+, max(case when label = 'TIDALVOLUME' then valuenum else null end) as tidalvolume
+, max(case when label = 'VENTILATIONRATE' then valuenum else null end) as ventilationrate
+, max(case when label = 'VENTILATOR' then valuenum else null end) as ventilator
+from pvt
+group by pvt.subject_id, pvt.hadm_id, pvt.icustay_id, pvt.charttime
+order by pvt.subject_id, pvt.hadm_id, pvt.icustay_id, pvt.charttime;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/22_blood_gas_first_day_arterial.sql b/mimic-iii-carevue/code/concepts/22_blood_gas_first_day_arterial.sql
new file mode 100644
index 000000000..5c3da6d71
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/22_blood_gas_first_day_arterial.sql
@@ -0,0 +1,140 @@
+drop table if exists blood_gas_first_day_arterial; create table blood_gas_first_day_arterial as
+
+with stg_spo2 as
+(
+ select subject_id, hadm_id, icustay_id, charttime
+ -- max here is just used to group SpO2 by charttime
+ , max(case when valuenum <= 0 or valuenum > 100 then null else valuenum end) as spo2
+ from chartevents
+ -- o2 sat
+ where itemid in (646) -- SpO2
+ group by subject_id, hadm_id, icustay_id, charttime
+)
+, stg_fio2 as
+(
+ select subject_id, hadm_id, icustay_id, charttime
+ -- pre-process the FiO2s to ensure they are between 21-100%
+ , max(
+ case
+ when itemid in (3420, 3422)
+ -- all these values are well formatted
+ then valuenum
+ when itemid = 190 and valuenum > 0.20 and valuenum < 1
+ -- well formatted but not in %
+ then valuenum * 100
+ else null end
+ ) as fio2_chartevents
+ from chartevents
+ where itemid in
+ (
+ 3420 -- FiO2
+ , 190 -- FiO2 set
+ , 3422 -- FiO2 [measured]
+ )
+ -- exclude rows marked as error
+ and (error is null or error = 0)
+ group by subject_id, hadm_id, icustay_id, charttime
+)
+, stg2 as
+(
+select bg.*
+ , row_number() over (partition by bg.icustay_id, bg.charttime order by s1.charttime desc) as lastrowspo2
+ , s1.spo2
+from blood_gas_first_day bg
+left join stg_spo2 s1
+ -- same patient
+ on bg.icustay_id = s1.icustay_id
+ -- spo2 occurred at most 2 hours before this blood gas
+ and s1.charttime >= (bg.charttime - interval '2 hour')
+ and s1.charttime <= bg.charttime
+where bg.po2 is not null
+)
+, stg3 as
+(
+select bg.*
+ , row_number() over (partition by bg.icustay_id, bg.charttime order by s2.charttime desc) as lastrowfio2
+ , s2.fio2_chartevents
+
+ -- create our specimen prediction
+ , 1/(1+exp(-(-0.02544
+ + 0.04598 * po2
+ + coalesce(-0.15356 * spo2 , -0.15356 * 97.49420 + 0.13429)
+ + coalesce( 0.00621 * fio2_chartevents , 0.00621 * 51.49550 + -0.24958)
+ + coalesce( 0.10559 * hemoglobin , 0.10559 * 10.32307 + 0.05954)
+ + coalesce( 0.13251 * so2 , 0.13251 * 93.66539 + -0.23172)
+ + coalesce(-0.01511 * pco2 , -0.01511 * 42.08866 + -0.01630)
+ + coalesce( 0.01480 * fio2 , 0.01480 * 63.97836 + -0.31142)
+ + coalesce(-0.00200 * aado2 , -0.00200 * 442.21186 + -0.01328)
+ + coalesce(-0.03220 * bicarbonate , -0.03220 * 22.96894 + -0.06535)
+ + coalesce( 0.05384 * totalco2 , 0.05384 * 24.72632 + -0.01405)
+ + coalesce( 0.08202 * lactate , 0.08202 * 3.06436 + 0.06038)
+ + coalesce( 0.10956 * ph , 0.10956 * 7.36233 + -0.00617)
+ + coalesce( 0.00848 * o2flow , 0.00848 * 7.59362 + -0.35803)
+ ))) as specimen_prob
+from stg2 bg
+left join stg_fio2 s2
+ -- same patient
+ on bg.icustay_id = s2.icustay_id
+ -- fio2 occurred at most 4 hours before this blood gas
+ and s2.charttime between (bg.charttime - interval '4 hour') and bg.charttime
+where bg.lastrowspo2 = 1 -- only the row with the most recent SpO2 (if no SpO2 found lastRowSpO2 = 1)
+)
+
+select subject_id, hadm_id,
+icustay_id, charttime
+, specimen -- raw data indicating sample type, only present 80% of the time
+
+-- prediction of specimen for missing data
+, case
+ when specimen is not null then specimen
+ when specimen_prob > 0.75 then 'ART'
+ else null end as specimen_pred
+, specimen_prob
+
+-- oxygen related parameters
+, so2, spo2 -- note spo2 is from chartevents
+, po2, pco2
+, fio2_chartevents, fio2
+, aado2
+-- also calculate AADO2
+, case
+ when PO2 is not null
+ and pco2 is not null
+ and coalesce(fio2, fio2_chartevents) is not null
+ -- multiple by 100 because FiO2 is in a % but should be a fraction
+ then (coalesce(fio2, fio2_chartevents)/100) * (760 - 47) - (pco2/0.8) - po2
+ else null
+ end as aado2_calc
+, case
+ when po2 is not null and coalesce(fio2, fio2_chartevents) is not null
+ -- multiply by 100 because FiO2 is in a % but should be a fraction
+ then 100*po2/(coalesce(fio2, fio2_chartevents))
+ else null
+ end as pao2fio2
+-- acid-base parameters
+, ph, baseexcess
+, bicarbonate, totalco2
+
+-- blood count parameters
+, hematocrit
+, hemoglobin
+, carboxyhemoglobin
+, methemoglobin
+
+-- chemistry
+, chloride, calcium
+, temperature
+, potassium, sodium
+, lactate
+, glucose
+
+-- ventilation stuff that's sometimes input
+, intubated, tidalvolume, ventilationrate, ventilator
+, peep, o2flow
+, requiredo2
+
+from stg3
+where lastrowfio2 = 1 -- only the most recent FiO2
+-- restrict it to *only* arterial samples
+and (specimen = 'ART' or specimen_prob > 0.75)
+order by icustay_id, charttime;
diff --git a/mimic-iii-carevue/code/concepts/23_gcs_first_day.sql b/mimic-iii-carevue/code/concepts/23_gcs_first_day.sql
new file mode 100644
index 000000000..48b4c68a6
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/23_gcs_first_day.sql
@@ -0,0 +1,129 @@
+drop table if exists gcs_first_day; create table gcs_first_day as
+-- itemids used:
+
+-- CAREVUE
+-- 723 as GCSVerbal
+-- 454 as GCSMotor
+-- 184 as GCSEyes
+
+
+-- The code combines the ITEMIDs into the carevue itemids, then pivots those
+-- So 223900 is changed to 723, then the ITEMID 723 is pivoted to form GCSVerbal
+
+-- Note:
+-- The GCS for sedated patients is defaulted to 15 in this code.
+-- This is in line with how the data is meant to be collected.
+-- e.g., from the SAPS II publication:
+-- For sedated patients, the Glasgow Coma Score before sedation was used.
+-- This was ascertained either from interviewing the physician who ordered the sedation,
+-- or by reviewing the patient's medical record.
+
+with base as
+(
+ select pvt.icustay_id
+ , pvt.charttime
+
+ -- Easier names - note we coalesced CareVue IDs below
+ , max(case when pvt.itemid = 454 then pvt.valuenum else null end) as gcsmotor
+ , max(case when pvt.itemid = 723 then pvt.valuenum else null end) as gcsverbal
+ , max(case when pvt.itemid = 184 then pvt.valuenum else null end) as gcseyes
+
+ -- If verbal was set to 0 in the below select, then this is an intubated patient
+ , case
+ when max(case when pvt.itemid = 723 then pvt.valuenum else null end) = 0
+ then 1
+ else 0
+ end as endotrachflag
+
+ , row_number ()
+ over (partition by pvt.icustay_id order by pvt.charttime asc) as rn
+
+ from (
+ select
+ l.icustay_id
+ , itemid
+
+ -- convert the data into a number, reserving a value of 0 for ET/Trach
+ , case
+ -- endotrach/vent is assigned a value of 0, later parsed specially
+ when l.itemid = 723 and l.value = '1.0 ET/Trach' then 0 -- carevue
+ else valuenum
+ end
+ as valuenum
+ , l.charttime
+ from chartevents l
+
+ -- get intime for charttime subselection
+ inner join icustays b
+ on l.icustay_id = b.icustay_id
+
+ -- Isolate the desired GCS variables
+ where l.itemid in
+ (
+ -- 198 -- GCS
+ -- GCS components, CareVue
+ 184, 454, 723
+ )
+ -- Only get data for the first 24 hours
+ and l.charttime between b.intime and (b.intime + interval '1 day')
+ -- exclude rows marked as error
+ and (l.error is null or l.error = 0)
+ ) pvt
+ group by pvt.icustay_id, pvt.charttime
+)
+, gcs as (
+ select b.*
+ , b2.gcsverbal as gcsverbalprev
+ , b2.gcsmotor as gcsmotorprev
+ , b2.gcseyes as gcseyesprev
+ -- Calculate GCS, factoring in special case when they are intubated and prev vals
+ -- note that the coalesce are used to implement the following if:
+ -- if current value exists, use it
+ -- if previous value exists, use it
+ -- otherwise, default to normal
+ , case
+ -- replace GCS during sedation with 15
+ when b.gcsverbal = 0
+ then 15
+ when b.gcsverbal is null and b2.gcsverbal = 0
+ then 15
+ -- if previously they were intub, but they aren't now, do not use previous GCS values
+ when b2.gcsverbal = 0
+ then
+ coalesce(b.gcsmotor,6)
+ + coalesce(b.gcsverbal,5)
+ + coalesce(b.gcseyes,4)
+ -- otherwise, add up score normally, imputing previous value if none available at current time
+ else
+ coalesce(b.gcsmotor,coalesce(b2.gcsmotor,6))
+ + coalesce(b.gcsverbal,coalesce(b2.gcsverbal,5))
+ + coalesce(b.gcseyes,coalesce(b2.gcseyes,4))
+ end as gcs
+
+ from base b
+ -- join to itself within 6 hours to get previous value
+ left join base b2
+ on b.icustay_id = b2.icustay_id and b.rn = b2.rn+1 and b2.charttime > (b.charttime - interval '6 hour')
+)
+, gcs_final as (
+ select gcs.*
+ -- This sorts the data by GCS, so rn=1 is the the lowest GCS values to keep
+ , row_number ()
+ over (partition by gcs.icustay_id
+ order by gcs.gcs
+ ) as ismingcs
+ from gcs
+)
+select ie.subject_id, ie.hadm_id, ie.icustay_id
+-- The minimum GCS is determined by the above row partition, we only join if IsMinGCS=1
+, gcs as mingcs
+, coalesce(gcsmotor,gcsmotorprev) as gcsmotor
+, coalesce(gcsverbal,gcsverbalprev) as gcsverbal
+, coalesce(gcseyes,gcseyesprev) as gcseyes
+, endotrachflag as endotrachflag
+
+-- subselect down to the cohort of eligible patients
+from icustays ie
+left join gcs_final gs
+ on ie.icustay_id = gs.icustay_id and gs.ismingcs = 1
+order by ie.icustay_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/24_height_first_day.sql b/mimic-iii-carevue/code/concepts/24_height_first_day.sql
new file mode 100644
index 000000000..85998bfb5
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/24_height_first_day.sql
@@ -0,0 +1,50 @@
+drop table if exists height_first_day; create table height_first_day as
+-- This query extracts heights for adult ICU patients.
+-- It uses all information from the patient's first ICU day.
+-- This is done for consistency with other queries - it's not necessarily needed.
+-- Height is unlikely to change throughout a patient's stay.
+-- staging table to ensure all heights are in centimeters
+with ce0 as
+(
+ select
+ c.icustay_id
+ , case
+ -- convert inches to centimetres
+ when itemid in (920, 1394, 4187, 3486)
+ then valuenum * 2.54
+ else valuenum
+ end as height
+ from chartevents c
+ inner join icustays ie
+ on c.icustay_id = ie.icustay_id
+ and c.charttime <= (ie.intime + interval '1 day')
+ and c.charttime > (ie.intime - interval '1 day') -- some fuzziness for admit time
+ where c.valuenum is not null
+ and c.itemid in (920, 1394, 4187, 3486, 3485, 4188) -- height
+ and c.valuenum != 0
+ -- exclude rows marked as error
+ and (c.error is null or c.error = 0)
+)
+, ce as
+(
+ select
+ icustay_id
+ -- extract the median height from the chart to add robustness against outliers
+ , avg(height) as height_chart
+ from ce0
+ where height > 100
+ group by icustay_id
+)
+
+select
+ ie.icustay_id
+ , ce.height_chart as height
+ -- components
+ , ce.height_chart
+from icustays ie
+-- filter to only adults
+inner join patients pat
+ on ie.subject_id = pat.subject_id
+ and ie.intime > (pat.dob + interval '1 year')
+left join ce
+ on ie.icustay_id = ce.icustay_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/25_labs_first_day.sql b/mimic-iii-carevue/code/concepts/25_labs_first_day.sql
new file mode 100644
index 000000000..0d6dbee01
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/25_labs_first_day.sql
@@ -0,0 +1,154 @@
+drop table if exists labs_first_day; create table labs_first_day as
+-- This query pivots lab values taken in the first 24 hours of a patient's stay
+
+-- Have already confirmed that the unit of measurement is always the same: null or the correct unit
+
+select
+ pvt.subject_id, pvt.hadm_id, pvt.icustay_id
+
+ , min(case when label = 'ANION GAP' then valuenum else null end) as aniongap_min
+ , max(case when label = 'ANION GAP' then valuenum else null end) as aniongap_max
+ , min(case when label = 'ALBUMIN' then valuenum else null end) as albumin_min
+ , max(case when label = 'ALBUMIN' then valuenum else null end) as albumin_max
+ , min(case when label = 'BANDS' then valuenum else null end) as bands_min
+ , max(case when label = 'BANDS' then valuenum else null end) as bands_max
+ , min(case when label = 'BICARBONATE' then valuenum else null end) as bicarbonate_min
+ , max(case when label = 'BICARBONATE' then valuenum else null end) as bicarbonate_max
+ , min(case when label = 'BILIRUBIN' then valuenum else null end) as bilirubin_min
+ , max(case when label = 'BILIRUBIN' then valuenum else null end) as bilirubin_max
+ , min(case when label = 'CREATININE' then valuenum else null end) as creatinine_min
+ , max(case when label = 'CREATININE' then valuenum else null end) as creatinine_max
+ , min(case when label = 'CHLORIDE' then valuenum else null end) as chloride_min
+ , max(case when label = 'CHLORIDE' then valuenum else null end) as chloride_max
+ , min(case when label = 'GLUCOSE' then valuenum else null end) as glucose_min
+ , max(case when label = 'GLUCOSE' then valuenum else null end) as glucose_max
+ , min(case when label = 'HEMATOCRIT' then valuenum else null end) as hematocrit_min
+ , max(case when label = 'HEMATOCRIT' then valuenum else null end) as hematocrit_max
+ , min(case when label = 'HEMOGLOBIN' then valuenum else null end) as hemoglobin_min
+ , max(case when label = 'HEMOGLOBIN' then valuenum else null end) as hemoglobin_max
+ , min(case when label = 'LACTATE' then valuenum else null end) as lactate_min
+ , max(case when label = 'LACTATE' then valuenum else null end) as lactate_max
+ , min(case when label = 'PLATELET' then valuenum else null end) as platelet_min
+ , max(case when label = 'PLATELET' then valuenum else null end) as platelet_max
+ , min(case when label = 'POTASSIUM' then valuenum else null end) as potassium_min
+ , max(case when label = 'POTASSIUM' then valuenum else null end) as potassium_max
+ , min(case when label = 'PTT' then valuenum else null end) as ptt_min
+ , max(case when label = 'PTT' then valuenum else null end) as ptt_max
+ , min(case when label = 'INR' then valuenum else null end) as inr_min
+ , max(case when label = 'INR' then valuenum else null end) as inr_max
+ , min(case when label = 'PT' then valuenum else null end) as pt_min
+ , max(case when label = 'PT' then valuenum else null end) as pt_max
+ , min(case when label = 'SODIUM' then valuenum else null end) as sodium_min
+ , max(case when label = 'SODIUM' then valuenum else null end) as sodium_max
+ , min(case when label = 'BUN' then valuenum else null end) as bun_min
+ , max(case when label = 'BUN' then valuenum else null end) as bun_max
+ , min(case when label = 'WBC' then valuenum else null end) as wbc_min
+ , max(case when label = 'WBC' then valuenum else null end) as wbc_max
+
+
+from
+( -- begin query that extracts the data
+ select ie.subject_id, ie.hadm_id, ie.icustay_id
+ -- here we assign labels to itemids
+ -- this also fuses together multiple itemids containing the same data
+ , case
+ when itemid = 50868 then 'ANION GAP'
+ when itemid = 50862 then 'ALBUMIN'
+ when itemid = 51144 then 'BANDS'
+ when itemid = 50882 then 'BICARBONATE'
+ when itemid = 50885 then 'BILIRUBIN'
+ when itemid = 50912 then 'CREATININE'
+ when itemid = 50806 then 'CHLORIDE'
+ when itemid = 50902 then 'CHLORIDE'
+ when itemid = 50809 then 'GLUCOSE'
+ when itemid = 50931 then 'GLUCOSE'
+ when itemid = 50810 then 'HEMATOCRIT'
+ when itemid = 51221 then 'HEMATOCRIT'
+ when itemid = 50811 then 'HEMOGLOBIN'
+ when itemid = 51222 then 'HEMOGLOBIN'
+ when itemid = 50813 then 'LACTATE'
+ when itemid = 51265 then 'PLATELET'
+ when itemid = 50822 then 'POTASSIUM'
+ when itemid = 50971 then 'POTASSIUM'
+ when itemid = 51275 then 'PTT'
+ when itemid = 51237 then 'INR'
+ when itemid = 51274 then 'PT'
+ when itemid = 50824 then 'SODIUM'
+ when itemid = 50983 then 'SODIUM'
+ when itemid = 51006 then 'BUN'
+ when itemid = 51300 then 'WBC'
+ when itemid = 51301 then 'WBC'
+ else null
+ end as label
+ , -- add in some sanity checks on the values
+ -- the where clause below requires all valuenum to be > 0, so these are only upper limit checks
+ case
+ when itemid = 50862 and valuenum > 10 then null -- g/dL 'ALBUMIN'
+ when itemid = 50868 and valuenum > 10000 then null -- mEq/L 'ANION GAP'
+ when itemid = 51144 and valuenum < 0 then null -- immature band forms, %
+ when itemid = 51144 and valuenum > 100 then null -- immature band forms, %
+ when itemid = 50882 and valuenum > 10000 then null -- mEq/L 'BICARBONATE'
+ when itemid = 50885 and valuenum > 150 then null -- mg/dL 'BILIRUBIN'
+ when itemid = 50806 and valuenum > 10000 then null -- mEq/L 'CHLORIDE'
+ when itemid = 50902 and valuenum > 10000 then null -- mEq/L 'CHLORIDE'
+ when itemid = 50912 and valuenum > 150 then null -- mg/dL 'CREATININE'
+ when itemid = 50809 and valuenum > 10000 then null -- mg/dL 'GLUCOSE'
+ when itemid = 50931 and valuenum > 10000 then null -- mg/dL 'GLUCOSE'
+ when itemid = 50810 and valuenum > 100 then null -- % 'HEMATOCRIT'
+ when itemid = 51221 and valuenum > 100 then null -- % 'HEMATOCRIT'
+ when itemid = 50811 and valuenum > 50 then null -- g/dL 'HEMOGLOBIN'
+ when itemid = 51222 and valuenum > 50 then null -- g/dL 'HEMOGLOBIN'
+ when itemid = 50813 and valuenum > 50 then null -- mmol/L 'LACTATE'
+ when itemid = 51265 and valuenum > 10000 then null -- K/uL 'PLATELET'
+ when itemid = 50822 and valuenum > 30 then null -- mEq/L 'POTASSIUM'
+ when itemid = 50971 and valuenum > 30 then null -- mEq/L 'POTASSIUM'
+ when itemid = 51275 and valuenum > 150 then null -- sec 'PTT'
+ when itemid = 51237 and valuenum > 50 then null -- 'INR'
+ when itemid = 51274 and valuenum > 150 then null -- sec 'PT'
+ when itemid = 50824 and valuenum > 200 then null -- mEq/L == mmol/L 'SODIUM'
+ when itemid = 50983 and valuenum > 200 then null -- mEq/L == mmol/L 'SODIUM'
+ when itemid = 51006 and valuenum > 300 then null -- 'BUN'
+ when itemid = 51300 and valuenum > 1000 then null -- 'WBC'
+ when itemid = 51301 and valuenum > 1000 then null -- 'WBC'
+ else le.valuenum
+ end as valuenum
+
+ from icustays ie
+
+ left join labevents le
+ on le.subject_id = ie.subject_id and le.hadm_id = ie.hadm_id
+ and le.charttime between (ie.intime - interval '6 hour') and (ie.intime + interval '1 day')
+ and le.itemid in
+ (
+ -- comment is: LABEL | CATEGORY | FLUID | NUMBER OF ROWS IN LABEVENTS
+ 50868, -- ANION GAP | CHEMISTRY | BLOOD | 769895
+ 50862, -- ALBUMIN | CHEMISTRY | BLOOD | 146697
+ 51144, -- BANDS - hematology
+ 50882, -- BICARBONATE | CHEMISTRY | BLOOD | 780733
+ 50885, -- BILIRUBIN, TOTAL | CHEMISTRY | BLOOD | 238277
+ 50912, -- CREATININE | CHEMISTRY | BLOOD | 797476
+ 50902, -- CHLORIDE | CHEMISTRY | BLOOD | 795568
+ 50806, -- CHLORIDE, WHOLE BLOOD | BLOOD GAS | BLOOD | 48187
+ 50931, -- GLUCOSE | CHEMISTRY | BLOOD | 748981
+ 50809, -- GLUCOSE | BLOOD GAS | BLOOD | 196734
+ 51221, -- HEMATOCRIT | HEMATOLOGY | BLOOD | 881846
+ 50810, -- HEMATOCRIT, CALCULATED | BLOOD GAS | BLOOD | 89715
+ 51222, -- HEMOGLOBIN | HEMATOLOGY | BLOOD | 752523
+ 50811, -- HEMOGLOBIN | BLOOD GAS | BLOOD | 89712
+ 50813, -- LACTATE | BLOOD GAS | BLOOD | 187124
+ 51265, -- PLATELET COUNT | HEMATOLOGY | BLOOD | 778444
+ 50971, -- POTASSIUM | CHEMISTRY | BLOOD | 845825
+ 50822, -- POTASSIUM, WHOLE BLOOD | BLOOD GAS | BLOOD | 192946
+ 51275, -- PTT | HEMATOLOGY | BLOOD | 474937
+ 51237, -- INR(PT) | HEMATOLOGY | BLOOD | 471183
+ 51274, -- PT | HEMATOLOGY | BLOOD | 469090
+ 50983, -- SODIUM | CHEMISTRY | BLOOD | 808489
+ 50824, -- SODIUM, WHOLE BLOOD | BLOOD GAS | BLOOD | 71503
+ 51006, -- UREA NITROGEN | CHEMISTRY | BLOOD | 791925
+ 51301, -- WHITE BLOOD CELLS | HEMATOLOGY | BLOOD | 753301
+ 51300 -- WBC COUNT | HEMATOLOGY | BLOOD | 2371
+ )
+ and valuenum is not null and valuenum > 0 -- lab values cannot be 0 and cannot be negative
+) pvt
+group by pvt.subject_id, pvt.hadm_id, pvt.icustay_id
+order by pvt.subject_id, pvt.hadm_id, pvt.icustay_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/26_rrt_first_day.sql b/mimic-iii-carevue/code/concepts/26_rrt_first_day.sql
new file mode 100644
index 000000000..4b8c5d6aa
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/26_rrt_first_day.sql
@@ -0,0 +1,85 @@
+drop table if exists rrt_first_day; create table rrt_first_day as
+-- determines if patients received any dialysis during their stay
+
+-- Some example aggregate queries which summarize the data here.
+-- This query estimates 6.7% of ICU patients received RRT.
+ -- select count(rrt.icustay_id) as numobs
+ -- , sum(rrt) as numrrt
+ -- , sum(case when rrt=1 then 1 else 0 end)*100.0 / count(rrt.icustay_id)
+ -- as percent_rrt
+ -- from rrt
+ -- inner join icustays ie on rrt.icustay_id = ie.icustay_id
+ -- inner join patients p
+ -- on rrt.subject_id = p.subject_id
+ -- and p.dob < ie.intime - interval '1' year
+ -- inner join admissions adm
+ -- on rrt.hadm_id = adm.hadm_id;
+
+-- This query estimates that 4.6% of first ICU stays received RRT.
+ -- select
+ -- count(rrt.icustay_id) as numobs
+ -- , sum(rrt) as numrrt
+ -- , sum(case when rrt=1 then 1 else 0 end)*100.0 / count(rrt.icustay_id)
+ -- as percent_rrt
+ -- from
+ -- (
+ -- select ie.icustay_id, rrt.rrt
+ -- , row_number() over (partition by ie.subject_id order by ie.intime) rn
+ -- from rrt
+ -- inner join icustays ie
+ -- on rrt.icustay_id = ie.icustay_id
+ -- inner join patients p
+ -- on rrt.subject_id = p.subject_id
+ -- and p.dob < ie.intime - interval '1' year
+ -- inner join admissions adm
+ -- on rrt.hadm_id = adm.hadm_id
+ -- ) rrt
+ -- where rn = 1;
+
+with cv as
+(
+ select ie.icustay_id
+ , max(
+ case
+ when ce.itemid in (152,148,149,146,147,151,150) and value is not null then 1
+ when ce.itemid in (229,235,241,247,253,259,265,271) and value = 'Dialysis Line' then 1
+ when ce.itemid = 582 and value in ('CAVH Start','CAVH D/C','CVVHD Start','CVVHD D/C','Hemodialysis st','Hemodialysis end') then 1
+ else 0 end
+ ) as rrt
+ from icustays ie
+ inner join chartevents ce
+ on ie.icustay_id = ce.icustay_id
+ and ce.itemid in
+ (
+ 152 -- "Dialysis Type";61449
+ ,148 -- "Dialysis Access Site";60335
+ ,149 -- "Dialysis Access Type";60030
+ ,146 -- "Dialysate Flow ml/hr";57445
+ ,147 -- "Dialysate Infusing";56605
+ ,151 -- "Dialysis Site Appear";37345
+ ,150 -- "Dialysis Machine";27472
+ ,229 -- INV Line#1 [Type]
+ ,235 -- INV Line#2 [Type]
+ ,241 -- INV Line#3 [Type]
+ ,247 -- INV Line#4 [Type]
+ ,253 -- INV Line#5 [Type]
+ ,259 -- INV Line#6 [Type]
+ ,265 -- INV Line#7 [Type]
+ ,271 -- INV Line#8 [Type]
+ ,582 -- Procedures
+ )
+ and ce.value is not null
+ and ce.charttime between ie.intime and (ie.intime + interval '1 day')
+ where ie.dbsource = 'carevue'
+ group by ie.icustay_id
+)
+
+select ie.subject_id, ie.hadm_id, ie.icustay_id
+ , case
+ when cv.rrt = 1 then 1
+ else 0
+ end as rrt
+from icustays ie
+left join cv
+ on ie.icustay_id = cv.icustay_id
+order by ie.icustay_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/27_urine_output_first_day.sql b/mimic-iii-carevue/code/concepts/27_urine_output_first_day.sql
new file mode 100644
index 000000000..05fc05666
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/27_urine_output_first_day.sql
@@ -0,0 +1,38 @@
+drop table if exists urine_output_first_day; create table urine_output_first_day as
+-- ------------------------------------------------------------------
+-- Purpose: Create a view of the urine output for each icustay_id over the first 24 hours.
+-- ------------------------------------------------------------------
+
+select
+ -- patient identifiers
+ ie.subject_id, ie.hadm_id, ie.icustay_id
+
+ -- volumes associated with urine output ITEMIDs
+ , sum(oe.value) as urineoutput
+from icustays ie
+-- join to the outputevents table to get urine output
+left join outputevents oe
+-- join on all patient identifiers
+on ie.subject_id = oe.subject_id and ie.hadm_id = oe.hadm_id and ie.icustay_id = oe.icustay_id
+-- and ensure the data occurs during the first day
+and oe.charttime between ie.intime and (ie.intime + interval '1 day') -- first icu day
+where itemid in
+(
+-- these are the most frequently occurring urine output observations in CareVue
+40055, -- "Urine Out Foley"
+43175, -- "Urine"
+40069, -- "Urine Out Void"
+40094, -- "Urine Out Condom Cath"
+40715, -- "Urine Out Suprapubic"
+40473, -- "Urine Out IleoConduit"
+40085, -- "Urine Out Incontinent"
+40057, -- "Urine Out Rt Nephrostomy"
+40056, -- "Urine Out Lt Nephrostomy"
+40405, -- "Urine Out Other"
+40428, -- "Urine Out Straight Cath"
+40086, -- "Urine Out Incontinent"
+40096, -- "Urine Out Ureteral Stent #1"
+40651 -- "Urine Out Ureteral Stent #2"
+)
+group by ie.subject_id, ie.hadm_id, ie.icustay_id
+order by ie.subject_id, ie.hadm_id, ie.icustay_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/28_ventilation_first_day.sql b/mimic-iii-carevue/code/concepts/28_ventilation_first_day.sql
new file mode 100644
index 000000000..c870b7c99
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/28_ventilation_first_day.sql
@@ -0,0 +1,25 @@
+drop table if exists ventilation_first_day; create table ventilation_first_day as
+-- Determines if a patient is ventilated on the first day of their ICU stay.
+-- Creates a table with the result.
+-- Requires the `durations_ventilation_durations` table, generated by ../durations_ventilation_durations.sql
+
+select
+ ie.subject_id, ie.hadm_id, ie.icustay_id
+ -- if vd.icustay_id is not null, then they have a valid ventilation event
+ -- in this case, we say they are ventilated
+ -- otherwise, they are not
+ , max(case
+ when vd.icustay_id is not null then 1
+ else 0 end) as vent
+from icustays ie
+left join ventilation_durations vd
+ on ie.icustay_id = vd.icustay_id
+ and
+ (
+ -- ventilation duration overlaps with ICU admission -> vented on admission
+ (vd.starttime <= ie.intime and vd.endtime >= ie.intime)
+ -- ventilation started during the first day
+ or (vd.starttime >= ie.intime and vd.starttime <= (ie.intime + interval '1 day'))
+ )
+group by ie.subject_id, ie.hadm_id, ie.icustay_id
+order by ie.subject_id, ie.hadm_id, ie.icustay_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/29_vitals_first_day.sql b/mimic-iii-carevue/code/concepts/29_vitals_first_day.sql
new file mode 100644
index 000000000..ddcdd59cd
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/29_vitals_first_day.sql
@@ -0,0 +1,98 @@
+drop table if exists vitals_first_day; create table vitals_first_day as
+-- This query pivots the vital signs for the first 24 hours of a patient's stay
+-- Vital signs include heart rate, blood pressure, respiration rate, and temperature
+
+select pvt.subject_id, pvt.hadm_id, pvt.icustay_id
+ -- easier names
+ , min(case when vitalid = 1 then valuenum else null end) as heartrate_min
+ , max(case when vitalid = 1 then valuenum else null end) as heartrate_max
+ , avg(case when vitalid = 1 then valuenum else null end) as heartrate_mean
+ , min(case when vitalid = 2 then valuenum else null end) as sysbp_min
+ , max(case when vitalid = 2 then valuenum else null end) as sysbp_max
+ , avg(case when vitalid = 2 then valuenum else null end) as sysbp_mean
+ , min(case when vitalid = 3 then valuenum else null end) as diasbp_min
+ , max(case when vitalid = 3 then valuenum else null end) as diasbp_max
+ , avg(case when vitalid = 3 then valuenum else null end) as diasbp_mean
+ , min(case when vitalid = 4 then valuenum else null end) as meanbp_min
+ , max(case when vitalid = 4 then valuenum else null end) as meanbp_max
+ , avg(case when vitalid = 4 then valuenum else null end) as meanbp_mean
+ , min(case when vitalid = 5 then valuenum else null end) as resprate_min
+ , max(case when vitalid = 5 then valuenum else null end) as resprate_max
+ , avg(case when vitalid = 5 then valuenum else null end) as resprate_mean
+ , min(case when vitalid = 6 then valuenum else null end) as tempc_min
+ , max(case when vitalid = 6 then valuenum else null end) as tempc_max
+ , avg(case when vitalid = 6 then valuenum else null end) as tempc_mean
+ , min(case when vitalid = 7 then valuenum else null end) as spo2_min
+ , max(case when vitalid = 7 then valuenum else null end) as spo2_max
+ , avg(case when vitalid = 7 then valuenum else null end) as spo2_mean
+ , min(case when vitalid = 8 then valuenum else null end) as glucose_min
+ , max(case when vitalid = 8 then valuenum else null end) as glucose_max
+ , avg(case when vitalid = 8 then valuenum else null end) as glucose_mean
+from
+ (
+ select
+ ie.subject_id, ie.hadm_id, ie.icustay_id
+ , case
+ when itemid in (211) and valuenum > 0 and valuenum < 300 then 1 -- HeartRate
+ when itemid in (51,442,455,6701) and valuenum > 0 and valuenum < 400 then 2 -- SysBP
+ when itemid in (8368,8440,8441,8555) and valuenum > 0 and valuenum < 300 then 3 -- DiasBP
+ when itemid in (456,52,6702,443) and valuenum > 0 and valuenum < 300 then 4 -- MeanBP
+ when itemid in (615,618) and valuenum > 0 and valuenum < 70 then 5 -- RespRate
+ when itemid in (678) and valuenum > 70 and valuenum < 120 then 6 -- TempF, converted to degC in valuenum call
+ when itemid in (676) and valuenum > 10 and valuenum < 50 then 6 -- TempC
+ when itemid in (646) and valuenum > 0 and valuenum <= 100 then 7 -- SpO2
+ when itemid in (807,811,1529,3745,3744) and valuenum > 0 then 8 -- Glucose
+ else null end as vitalid
+ -- convert F to C
+ , case when itemid in (678) then (valuenum-32)/1.8 else valuenum end as valuenum
+ from icustays ie
+ left join chartevents ce
+ on ie.icustay_id = ce.icustay_id
+ and ce.charttime between ie.intime and (ie.intime + interval '1 day')
+ and (cast(extract(epoch from (ce.charttime - ie.intime)) as numeric)) > 0
+ and (cast(extract(epoch from (ie.outtime - ie.intime))/(60*60) as numeric)) <= 24
+ -- exclude rows marked as error
+ and (ce.error is null or ce.error = 0)
+ where ce.itemid in
+ (
+ -- HEART RATE
+ 211, --"Heart Rate"
+
+ -- Systolic/diastolic
+ 51, -- Arterial BP [Systolic]
+ 442, -- Manual BP [Systolic]
+ 455, -- NBP [Systolic]
+ 6701, -- Arterial BP #2 [Systolic]
+
+ 8368, -- Arterial BP [Diastolic]
+ 8440, -- Manual BP [Diastolic]
+ 8441, -- NBP [Diastolic]
+ 8555, -- Arterial BP #2 [Diastolic]
+
+ -- MEAN ARTERIAL PRESSURE
+ 456, --"NBP Mean"
+ 52, --"Arterial BP Mean"
+ 6702, -- Arterial BP Mean #2
+ 443, -- Manual BP Mean(calc)
+
+ -- RESPIRATORY RATE
+ 618,-- Respiratory Rate
+ 615,-- Resp Rate (Total)
+
+ -- SPO2, peripheral
+ 646,
+
+ -- GLUCOSE, both lab and fingerstick
+ 807,-- Fingerstick Glucose
+ 811,-- Glucose (70-105)
+ 1529,-- Glucose
+ 3745,-- BloodGlucose
+ 3744,-- Blood Glucose
+
+ -- TEMPERATURE
+ 676, -- "Temperature C"
+ 678 -- "Temperature F"
+ )
+ ) pvt
+group by pvt.subject_id, pvt.hadm_id, pvt.icustay_id
+order by pvt.subject_id, pvt.hadm_id, pvt.icustay_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/30_weight_first_day.sql b/mimic-iii-carevue/code/concepts/30_weight_first_day.sql
new file mode 100644
index 000000000..7ad05e88a
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/30_weight_first_day.sql
@@ -0,0 +1,67 @@
+drop table if exists weight_first_day; create table weight_first_day as
+-- This query extracts weights for adult ICU patients on their first ICU day.
+-- It does *not* use any information after the first ICU day, as weight is
+-- sometimes used to monitor fluid balance.
+
+-- ** Requires the echodata view, generated by concepts/echo-data.sql
+
+with ce as
+(
+ select
+ c.icustay_id
+ -- we take the avg value from roughly first day
+ -- TODO: eliminate obvious outliers if there is a reasonable weight
+ -- (e.g. weight of 180kg and 90kg would remove 180kg instead of taking the median)
+ , avg(valuenum) as weight_admit
+ from chartevents c
+ inner join icustays ie
+ on c.icustay_id = ie.icustay_id
+ and c.charttime <= (ie.intime + interval '1 day')
+ and c.charttime > (ie.intime - interval '1 day') -- some fuzziness for admit time
+ where c.valuenum is not null
+ and c.itemid in (762) -- admit wt
+ and c.valuenum != 0
+ -- exclude rows marked as error
+ and (c.error is null or c.error = 0)
+ group by c.icustay_id
+)
+, dwt as
+(
+ select
+ c.icustay_id
+ , avg(valuenum) as weight_daily
+ from chartevents c
+ inner join icustays ie
+ on c.icustay_id = ie.icustay_id
+ and c.charttime <= (ie.intime + interval '1 day')
+ and c.charttime > (ie.intime - interval '1 day') -- some fuzziness for admit time
+ where c.valuenum is not null
+ and c.itemid in (763) -- daily weight
+ and c.valuenum != 0
+ -- exclude rows marked as error
+ and (c.error is null or c.error = 0)
+ group by c.icustay_id
+)
+
+select
+ ie.icustay_id
+ , round(cast(case
+ when ce.icustay_id is not null then ce.weight_admit
+ when dwt.icustay_id is not null then dwt.weight_daily
+ else null end as numeric), 2)
+ as weight
+ -- components
+ , ce.weight_admit
+ , dwt.weight_daily
+from icustays ie
+-- filter to only adults
+inner join patients pat
+ on ie.subject_id = pat.subject_id
+ and ie.intime > (pat.dob + interval '1 year')
+-- admission weight
+left join ce
+ on ie.icustay_id = ce.icustay_id
+-- daily weights
+left join dwt
+ on ie.icustay_id = dwt.icustay_id
+order by ie.icustay_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/31_urine_output.sql b/mimic-iii-carevue/code/concepts/31_urine_output.sql
new file mode 100644
index 000000000..58c8f375b
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/31_urine_output.sql
@@ -0,0 +1,28 @@
+drop table if exists urine_output; create table urine_output as
+-- First we drop the table if it exists
+select
+ oe.icustay_id
+ , oe.charttime
+ , sum(oe.value) as value
+from outputevents oe
+where oe.itemid in
+(
+ -- these are the most frequently occurring urine output observations in CareVue
+ 40055, -- "Urine Out Foley"
+ 43175, -- "Urine"
+ 40069, -- "Urine Out Void"
+ 40094, -- "Urine Out Condom Cath"
+ 40715, -- "Urine Out Suprapubic"
+ 40473, -- "Urine Out IleoConduit"
+ 40085, -- "Urine Out Incontinent"
+ 40057, -- "Urine Out Rt Nephrostomy"
+ 40056, -- "Urine Out Lt Nephrostomy"
+ 40405, -- "Urine Out Other"
+ 40428, -- "Urine Out Straight Cath"
+ 40086, -- Urine Out Incontinent
+ 40096, -- "Urine Out Ureteral Stent #1"
+ 40651 -- "Urine Out Ureteral Stent #2"
+)
+and oe.value < 5000 -- sanity check on urine value
+and oe.icustay_id is not null
+group by icustay_id, charttime;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/32_angus.sql b/mimic-iii-carevue/code/concepts/32_angus.sql
new file mode 100644
index 000000000..b9c06c542
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/32_angus.sql
@@ -0,0 +1,103 @@
+drop table if exists angus; create table angus as
+-- ICD-9 codes for Angus criteria of sepsis
+
+-- Angus et al, 2001. Epidemiology of severe sepsis in the United States
+-- http://www.ncbi.nlm.nih.gov/pubmed/11445675
+
+-- Case selection and definitions
+-- To identify cases with severe sepsis, we selected all acute care
+-- hospitalizations with ICD-9-CM codes for both:
+-- (a) a bacterial or fungal infectious process AND
+-- (b) a diagnosis of acute organ dysfunction (Appendix 2).
+
+-- ICD-9 codes for infection - as sourced from Appendix 1 of above paper
+with infection_group as
+(
+ select subject_id, hadm_id,
+ case
+ when substr(icd9_code,1,3) in ('001','002','003','004','005','008',
+ '009','010','011','012','013','014','015','016','017','018',
+ '020','021','022','023','024','025','026','027','030','031',
+ '032','033','034','035','036','037','038','039','040','041',
+ '090','091','092','093','094','095','096','097','098','100',
+ '101','102','103','104','110','111','112','114','115','116',
+ '117','118','320','322','324','325','420','421','451','461',
+ '462','463','464','465','481','482','485','486','494','510',
+ '513','540','541','542','566','567','590','597','601','614',
+ '615','616','681','682','683','686','730') THEN 1
+ when substr(icd9_code,1,4) in ('5695','5720','5721','5750','5990','7110',
+ '7907','9966','9985','9993') then 1
+ when substr(icd9_code,1,5) in ('49121','56201','56203','56211','56213',
+ '56983') then 1
+ else 0 end as infection
+ from diagnoses_icd
+),
+-- ICD-9 codes for organ dysfunction - as sourced from Appendix 2 of above paper
+organ_diag_group as
+(
+ select subject_id, hadm_id,
+ case
+ -- acute organ dysfunction diagnosis codes
+ when substr(icd9_code,1,3) in ('458','293','570','584') then 1
+ when substr(icd9_code,1,4) in ('7855','3483','3481',
+ '2874','2875','2869','2866','5734') then 1
+ else 0 end as organ_dysfunction,
+ -- Explicit diagnosis of severe sepsis or septic shock
+ case
+ when substr(icd9_code,1,5) in ('99592','78552') then 1
+ else 0 end as explicit_sepsis
+ from diagnoses_icd
+),
+-- Mechanical ventilation
+organ_proc_group as
+(
+ select subject_id, hadm_id,
+ case
+ when icd9_code in ('9670', '9671', '9672') then 1
+ else 0 end as mech_vent
+ from procedures_icd
+),
+-- Aggregate above views together
+aggregate as
+(
+ select subject_id, hadm_id,
+ case
+ when hadm_id in
+ (select distinct hadm_id
+ from infection_group
+ where infection = 1)
+ then 1
+ else 0 end as infection,
+ case
+ when hadm_id in
+ (select distinct hadm_id
+ from organ_diag_group
+ where explicit_sepsis = 1)
+ then 1
+ else 0 end as explicit_sepsis,
+ case
+ when hadm_id in
+ (select distinct hadm_id
+ from organ_diag_group
+ where organ_dysfunction = 1)
+ then 1
+ else 0 end as organ_dysfunction,
+ case
+ when hadm_id in
+ (select distinct hadm_id
+ from organ_proc_group
+ where mech_vent = 1)
+ then 1
+ else 0 end as mech_vent
+ from admissions
+)
+-- output component flags (explicit sepsis, organ dysfunction) and final flag (angus)
+select subject_id, hadm_id, infection,
+ explicit_sepsis, organ_dysfunction, mech_vent,
+case
+ when explicit_sepsis = 1 then 1
+ when infection = 1 and organ_dysfunction = 1 then 1
+ when infection = 1 and mech_vent = 1 then 1
+ else 0 end
+as angus
+from aggregate;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/33_martin.sql b/mimic-iii-carevue/code/concepts/33_martin.sql
new file mode 100644
index 000000000..20f53ffef
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/33_martin.sql
@@ -0,0 +1,111 @@
+drop table if exists martin; create table martin as
+-- ICD-9 codes for sepsis as validated by Martin et al.
+
+-- Greg S. Martin, David M. Mannino, Stephanie Eaton, and Marc Moss. The epidemiology of
+-- sepsis in the united states from 1979 through 2000. N Engl J Med, 348(16):1546–1554, Apr
+-- 2003. doi: 10.1056/NEJMoa022139. URL http://dx.doi.org/10.1056/NEJMoa022139.
+
+with co_dx as
+(
+ select subject_id, hadm_id
+ , max(
+ case
+ -- septicemia
+ when substr(icd9_code,1,3) = '038' then 1
+ -- septicemic, bacteremia, disseminated fungal infection, disseminated candida infection
+ -- NOTE: the paper specifies 020.0 ... but this is bubonic plague
+ -- presumably, they meant 020.2, which is septicemic plague
+ when substr(icd9_code,1,4) in ('0202','7907','1179','1125') then 1
+ -- disseminated fungal endocarditis
+ when substr(icd9_code,1,5) = '11281' then 1
+ else 0 end
+ ) as sepsis
+ , max(
+ case
+ when substr(icd9_code,1,4) in ('7991') then 1
+ when substr(icd9_code,1,5) in ('51881','51882','51885','78609') then 1
+ else 0 end
+ ) as respiratory
+ , max(
+ case
+ when substr(icd9_code,1,4) in ('4580','7855','4580','4588','4589','7963') then 1
+ when substr(icd9_code,1,5) in ('785.51','785.59') then 1
+ else 0 end
+ ) as cardiovascular
+ , max(
+ case
+ when substr(icd9_code,1,3) in ('584','580','585') then 1
+ else 0 end
+ ) as renal
+ , max(
+ case
+ when substr(icd9_code,1,3) in ('570') then 1
+ when substr(icd9_code,1,4) in ('5722','5733') then 1
+ else 0 end
+ ) as hepatic
+ , max(
+ case
+ when substr(icd9_code,1,4) in ('2862','2866','2869','2873','2874','2875') then 1
+ else 0 end
+ ) as hematologic
+ , max(
+ case
+ when substr(icd9_code,1,4) in ('2762') then 1
+ else 0 end
+ ) as metabolic
+ , max(
+ case
+ when substr(icd9_code,1,3) in ('293') then 1
+ when substr(icd9_code,1,4) in ('3481','3483') then 1
+ when substr(icd9_code,1,5) in ('78001','78009') then 1
+ else 0 end
+ ) as neurologic
+ from diagnoses_icd
+ group by subject_id, hadm_id
+)
+-- procedure codes:
+-- "96.7 - Ventilator management"
+-- translated:
+-- 9670 Continuous invasive mechanical ventilation of unspecified duration
+-- 9671 Continuous invasive mechanical ventilation for less than 96 consecutive hours
+-- 9672 Continuous invasive mechanical ventilation for 96 consecutive hours or more
+-- "39.95 - Hemodialysis"
+-- 3995 Hemodialysis
+-- "89.14 - Electroencephalography"
+-- 8914 Electroencephalogram
+, co_proc as
+(
+ select
+ subject_id, hadm_id
+ , max(case when icd9_code = '967' then 1 else 0 end) as respiratory
+ , max(case when icd9_code = '3995' then 1 else 0 end) as renal
+ , max(case when icd9_code = '8914' then 1 else 0 end) as neurologic
+ from procedures_icd
+ group by subject_id, hadm_id
+)
+select
+ adm.subject_id, adm.hadm_id
+ , co_dx.sepsis
+ , case
+ when co_dx.respiratory = 1 or co_proc.respiratory = 1
+ or co_dx.cardiovascular = 1
+ or co_dx.renal = 1 or co_proc.renal = 1
+ or co_dx.hepatic = 1
+ or co_dx.hematologic = 1
+ or co_dx.metabolic = 1
+ or co_dx.neurologic = 1 or co_proc.neurologic = 1
+ then 1
+ else 0
+ end as organ_failure
+ , case when co_dx.respiratory = 1 or co_proc.respiratory = 1 then 1 else 0 end as respiratory
+ , co_dx.cardiovascular
+ , case when co_dx.renal = 1 or co_proc.renal = 1 then 1 else 0 end as renal
+ , co_dx.hepatic
+ , co_dx.hematologic
+ , co_dx.metabolic
+ , case when co_dx.neurologic = 1 or co_proc.neurologic = 1 then 1 else 0 end as neurologic
+from admissions adm
+left join co_dx
+ on adm.hadm_id = co_dx.hadm_id
+left join co_proc
+ on adm.hadm_id = co_proc.hadm_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/34_explicit.sql b/mimic-iii-carevue/code/concepts/34_explicit.sql
new file mode 100644
index 000000000..3f851af87
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/34_explicit.sql
@@ -0,0 +1,35 @@
+drop table if exists explicit; create table explicit as
+-- This code extracts explicit sepsis using ICD-9 diagnosis codes
+-- That is, the two codes 995.92 (severe sepsis) or 785.52 (septic shock)
+-- These codes are extremely specific to sepsis, but have very low sensitivity
+-- From Iwashyna et al. (vs. chart reviews): 100% PPV, 9.3% sens, 100% specificity
+
+with co_dx as
+(
+ select hadm_id
+ -- sepsis codes
+ , max(
+ case
+ when icd9_code = '99592' then 1
+ else 0 end
+ ) as severe_sepsis
+ , max(
+ case
+ when icd9_code = '78552' then 1
+ else 0 end
+ ) as septic_shock
+ from diagnoses_icd
+ group by hadm_id
+)
+select
+ adm.subject_id
+ , adm.hadm_id
+ , co_dx.severe_sepsis
+ , co_dx.septic_shock
+ , case when co_dx.severe_sepsis = 1 or co_dx.septic_shock = 1
+ then 1
+ else 0 end as sepsis
+from admissions adm
+left join co_dx
+ on adm.hadm_id = co_dx.hadm_id
+order by adm.subject_id, adm.hadm_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/35_ccs_dx.sql b/mimic-iii-carevue/code/concepts/35_ccs_dx.sql
new file mode 100644
index 000000000..c5d34562c
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/35_ccs_dx.sql
@@ -0,0 +1,55 @@
+drop table if exists ccs_dx;
+create table ccs_dx
+(
+ icd9_code char(5) not null,
+ -- we will populate ccs_mid and ccs_name with the most granular id/name later
+ ccs_matched_id varchar(10),
+ ccs_matched_name varchar(100),
+ ccs_id integer,
+ ccs_name varchar(100),
+ -- ccs levels and names based on position in hierarchy
+ ccs_level1 varchar(10),
+ ccs_group1 varchar(100),
+ ccs_level2 varchar(10),
+ ccs_group2 varchar(100),
+ ccs_level3 varchar(10),
+ ccs_group3 varchar(100),
+ ccs_level4 varchar(10),
+ ccs_group4 varchar(100)
+);
+
+-- copy all columns *but* ccs_mid and ccs_name
+-- we will populate these after the data is loaded in
+\copy ccs_dx (icd9_code, ccs_level1, ccs_group1, ccs_level2, ccs_group2, ccs_level3, ccs_group3, ccs_level4, ccs_group4) from program 'gzip -dc ./code/concepts/dx_data/ccs_multi_dx.csv.gz' csv header;
+
+-- add in matched ID, name, and ccs_id
+-- matched id (ccs_mid): the ccs ID with the hierachy, e.g. 7.1.2.1
+-- name (ccs_name): the most granular CCS category the diagnosis is in
+-- ID (ccs_id): the CCS identifier for the ICD-9 code (integer)
+update ccs_dx
+set ccs_matched_id=tt.ccs_matched_id, ccs_matched_name=tt.ccs_matched_name,
+ ccs_name=tt.ccs_name, ccs_id=cast(tt.ccs_id as integer)
+from (
+ select icd9_code
+ , coalesce(ccs_level4, ccs_level3, ccs_level2, ccs_level1) as ccs_matched_id
+ -- remove the trailing ccs_id from name column, i.e. "Burns [240.]" -> "Burns"
+ , regexp_replace(coalesce(ccs_group4, ccs_group3, ccs_group2, ccs_group1), '\[[0-9]+\.\]$', '') as ccs_matched_name
+ -- ccs_id is sometimes present at a higher level of granularity
+ -- e.g. for 7.1.2.1, the CCS name is at level 7.1.2
+ -- therefore we pull from the first category to have the CCS ID
+ , case
+ when ccs_group4 ~ '\[([0-9]+)\.\]$' then regexp_replace(ccs_group4, '\[[0-9]+\.\]$', '')
+ when ccs_group3 ~ '\[([0-9]+)\.\]$' then regexp_replace(ccs_group3, '\[[0-9]+\.\]$', '')
+ when ccs_group2 ~ '\[([0-9]+)\.\]$' then regexp_replace(ccs_group2, '\[[0-9]+\.\]$', '')
+ when ccs_group1 ~ '\[([0-9]+)\.\]$' then regexp_replace(ccs_group1, '\[[0-9]+\.\]$', '')
+ else null end as ccs_name
+ -- extract the trailing ccs_id from name, i.e. "burns [240.]" -> "240"
+ , coalesce(
+ substring(ccs_group4, '\[([0-9]+)\.\]$'),
+ substring(ccs_group3, '\[([0-9]+)\.\]$'),
+ substring(ccs_group2, '\[([0-9]+)\.\]$'),
+ substring(ccs_group1, '\[([0-9]+)\.\]$')
+ ) as ccs_id
+ from ccs_dx
+) as tt
+where ccs_dx.icd9_code = tt.icd9_code;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/36_kdigo_creatinine.sql b/mimic-iii-carevue/code/concepts/36_kdigo_creatinine.sql
new file mode 100644
index 000000000..0df6a5cdf
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/36_kdigo_creatinine.sql
@@ -0,0 +1,38 @@
+drop table if exists kdigo_creatinine; create table kdigo_creatinine as
+-- Extract all creatinine values from labevents around patient's ICU stay
+with cr as
+(
+select
+ ie.icustay_id
+ , ie.intime, ie.outtime
+ , le.valuenum as creat
+ , le.charttime
+ from icustays ie
+ left join labevents le
+ on ie.subject_id = le.subject_id
+ and le.itemid = 50912
+ and le.valuenum is not null
+ and (cast(extract(epoch from (le.charttime - ie.intime))/(60*60) as numeric)) <= (7*24-6)
+ and le.charttime >= (ie.intime - interval '6 hour')
+ and le.charttime <= (ie.intime + interval '7 day')
+)
+-- add in the lowest value in the previous 48 hours/7 days
+select
+ cr.icustay_id
+ , cr.charttime
+ , cr.creat
+ , min(cr48.creat) as creat_low_past_48hr
+ , min(cr7.creat) as creat_low_past_7day
+from cr
+-- add in all creatinine values in the last 48 hours
+left join cr cr48
+ on cr.icustay_id = cr48.icustay_id
+ and cr48.charttime < cr.charttime
+ and (cast(extract(epoch from (cr.charttime - cr48.charttime))/(60*60) as numeric)) <= 48
+-- add in all creatinine values in the last 7 days
+left join cr cr7
+ on cr.icustay_id = cr7.icustay_id
+ and cr7.charttime < cr.charttime
+ and (cast(extract(epoch from (cr.charttime - cr7.charttime))/(60*60*24) as numeric)) <= 7
+group by cr.icustay_id, cr.charttime, cr.creat
+order by cr.icustay_id, cr.charttime, cr.creat;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/37_kdigo_uo.sql b/mimic-iii-carevue/code/concepts/37_kdigo_uo.sql
new file mode 100644
index 000000000..34eeaf622
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/37_kdigo_uo.sql
@@ -0,0 +1,83 @@
+drop table if exists kdigo_uo; create table kdigo_uo as
+with ur_stg as
+(
+ select io.icustay_id, io.charttime
+ -- we have joined each row to all rows preceding within 24 hours
+ -- we can now sum these rows to get total UO over the last 24 hours
+ -- we can use case statements to restrict it to only the last 6/12 hours
+ -- therefore we have three sums:
+ -- 1) over a 6 hour period
+ -- 2) over a 12 hour period
+ -- 3) over a 24 hour period
+
+ -- note that we assume data charted at charttime corresponds to 1 hour of UO
+ -- therefore we use '5' and '11' to restrict the period, rather than 6/12
+ -- this assumption may overestimate UO rate when documentation is done less than hourly
+ , sum(case when (cast(extract(epoch from (io.charttime - iosum.charttime))/(60*60) as numeric)) <= 5
+ then iosum.value
+ else null end) as urineoutput_6hr
+ , sum(case when (cast(extract(epoch from (io.charttime - iosum.charttime))/(60*60) as numeric)) <= 11
+ then iosum.value
+ else null end) as urineoutput_12hr
+ -- 24 hours
+ , sum(iosum.value) as urineoutput_24hr
+
+ -- retain the earliest time used for each summation
+ -- this is later used to tabulate rates
+ , min(case when io.charttime <= (iosum.charttime + interval '5 hour')
+ then iosum.charttime
+ else null end)
+ as starttime_6hr
+ , min(case when io.charttime <= (iosum.charttime + interval '11 hour')
+ then iosum.charttime
+ else null end)
+ as starttime_12hr
+ , min(iosum.charttime) as starttime_24hr
+ from urine_output io
+ -- this join gives you all UO measurements over a 24 hour period
+ left join urine_output iosum
+ on io.icustay_id = iosum.icustay_id
+ and io.charttime >= iosum.charttime
+ and io.charttime <= (iosum.charttime + interval '23 hour')
+ group by io.icustay_id, io.charttime
+)
+-- calculate hours used to sum UO over
+, ur_stg2 as
+(
+ select
+ icustay_id
+ , charttime
+ , urineoutput_6hr
+ , urineoutput_12hr
+ , urineoutput_24hr
+ -- calculate time over which we summed UO
+ -- note: adding 1 hour as we assume data charted corresponds to previous hour
+ -- i.e. if documentation is:
+ -- 10:00, 100 mL
+ -- 11:00, 50 mL
+ -- then this is two hours of documentation, even though (11:00 - 10:00) is 1 hour
+ , round((cast(extract(epoch from (charttime - starttime_6hr))/(60*60) as numeric)), 4) + 1 as uo_tm_6hr
+ , round((cast(extract(epoch from (charttime - starttime_12hr))/(60*60) as numeric)), 4) + 1 as uo_tm_12hr
+ , round((cast(extract(epoch from (charttime - starttime_24hr))/(60*60) as numeric)), 4) + 1 as uo_tm_24hr
+ from ur_stg
+)
+select
+ ur.icustay_id
+, ur.charttime
+, wd.weight
+, ur.urineoutput_6hr
+, ur.urineoutput_12hr
+, ur.urineoutput_24hr
+, round(cast((ur.urineoutput_6hr/wd.weight/uo_tm_6hr) as numeric), 4) as uo_rt_6hr
+, round(cast((ur.urineoutput_12hr/wd.weight/uo_tm_12hr) as numeric), 4) as uo_rt_12hr
+, round(cast((ur.urineoutput_24hr/wd.weight/uo_tm_24hr) as numeric), 4) as uo_rt_24hr
+-- time of earliest uo measurement that was used to calculate the rate
+, uo_tm_6hr
+, uo_tm_12hr
+, uo_tm_24hr
+from ur_stg2 ur
+left join weight_durations wd
+ on ur.icustay_id = wd.icustay_id
+ and ur.charttime >= wd.starttime
+ and ur.charttime < wd.endtime
+order by icustay_id, charttime;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/38_kdigo_stages.sql b/mimic-iii-carevue/code/concepts/38_kdigo_stages.sql
new file mode 100644
index 000000000..f9e00712d
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/38_kdigo_stages.sql
@@ -0,0 +1,91 @@
+drop table if exists kdigo_stages; create table kdigo_stages as
+-- This query checks if the patient had AKI according to KDIGO.
+-- AKI is calculated every time a creatinine or urine output measurement occurs.
+-- Baseline creatinine is defined as the lowest creatinine in the past 7 days.
+
+-- get creatinine stages
+with cr_stg as
+(
+ select
+ cr.icustay_id
+ , cr.charttime
+ , cr.creat
+ , case
+ -- 3x baseline
+ when cr.creat >= (cr.creat_low_past_7day*3.0) then 3
+ -- *OR* cr >= 4.0 with associated increase
+ when cr.creat >= 4
+ -- For patients reaching Stage 3 by SCr >4.0 mg/dl
+ -- require that the patient first achieve ... acute increase >= 0.3 within 48 hr
+ -- *or* an increase of >= 1.5 times baseline
+ and (cr.creat_low_past_48hr <= 3.7 or cr.creat >= (1.5*cr.creat_low_past_7day))
+ then 3
+ -- TODO: initiation of RRT
+ when cr.creat >= (cr.creat_low_past_7day*2.0) then 2
+ when cr.creat >= (cr.creat_low_past_48hr+0.3) then 1
+ when cr.creat >= (cr.creat_low_past_7day*1.5) then 1
+ else 0 end as aki_stage_creat
+ from kdigo_creatinine cr
+)
+-- stages for UO / creat
+, uo_stg as
+(
+ select
+ uo.icustay_id
+ , uo.charttime
+ , uo.weight
+ , uo.uo_rt_6hr
+ , uo.uo_rt_12hr
+ , uo.uo_rt_24hr
+ -- AKI stages according to urine output
+ , case
+ when uo.uo_rt_6hr is null then null
+ -- require patient to be in icu for at least 6 hours to stage uo
+ when uo.charttime <= (ie.intime + interval '6 hour') then 0
+ -- require the uo rate to be calculated over half the period
+ -- i.e. for uo rate over 24 hours, require documentation at least 12 hr apart
+ when uo.uo_tm_24hr >= 11 and uo.uo_rt_24hr < 0.3 then 3
+ when uo.uo_tm_12hr >= 5 and uo.uo_rt_12hr = 0 then 3
+ when uo.uo_tm_12hr >= 5 and uo.uo_rt_12hr < 0.5 then 2
+ when uo.uo_tm_6hr >= 2 and uo.uo_rt_6hr < 0.5 then 1
+ else 0 end as aki_stage_uo
+ from kdigo_uo uo
+ inner join icustays ie
+ on uo.icustay_id = ie.icustay_id
+)
+-- get all charttimes documented
+, tm_stg as
+(
+ select
+ icustay_id, charttime
+ from cr_stg
+ union distinct
+ select
+ icustay_id, charttime
+ from uo_stg
+)
+select
+ ie.icustay_id
+ , tm.charttime
+ , cr.creat
+ , cr.aki_stage_creat
+ , uo.uo_rt_6hr
+ , uo.uo_rt_12hr
+ , uo.uo_rt_24hr
+ , uo.aki_stage_uo
+ -- Classify AKI using both creatinine/urine output criteria
+ , greatest(
+ coalesce(cr.aki_stage_creat, 0),
+ coalesce(uo.aki_stage_uo, 0)
+ ) as aki_stage
+from icustays ie
+-- get all possible charttimes as listed in tm_stg
+left join tm_stg tm
+ on ie.icustay_id = tm.icustay_id
+left join cr_stg cr
+ on ie.icustay_id = cr.icustay_id
+ and tm.charttime = cr.charttime
+left join uo_stg uo
+ on ie.icustay_id = uo.icustay_id
+ and tm.charttime = uo.charttime
+order by ie.icustay_id, tm.charttime;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/39_kdigo_stages_7day.sql b/mimic-iii-carevue/code/concepts/39_kdigo_stages_7day.sql
new file mode 100644
index 000000000..4bff009f1
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/39_kdigo_stages_7day.sql
@@ -0,0 +1,68 @@
+drop table if exists kdigo_stages_7day; create table kdigo_stages_7day as
+-- This query checks if the patient had AKI during the first 7 days of their ICU
+-- stay according to the KDIGO guideline.
+-- https://kdigo.org/wp-content/uploads/2016/10/KDIGO-2012-AKI-Guideline-English.pdf
+
+-- get the worst staging of creatinine in the first 48 hours
+with cr_aki as
+(
+ select
+ k.icustay_id
+ , k.charttime
+ , k.creat
+ , k.aki_stage_creat
+ , row_number() over (partition by k.icustay_id order by k.aki_stage_creat desc, k.creat desc) as rn
+ from icustays ie
+ inner join kdigo_stages k
+ on ie.icustay_id = k.icustay_id
+ where (cast(extract(epoch from (k.charttime - ie.intime))/(60*60) as numeric)) > -6
+ and (cast(extract(epoch from (k.charttime - ie.intime))/(60*60*24) as numeric)) <= 7
+ and k.aki_stage_creat is not null
+)
+-- get the worst staging of urine output in the first 48 hours
+, uo_aki as
+(
+ select
+ k.icustay_id
+ , k.charttime
+ , k.uo_rt_6hr, k.uo_rt_12hr, k.uo_rt_24hr
+ , k.aki_stage_uo
+ , row_number() over
+ (
+ partition by k.icustay_id
+ order by k.aki_stage_uo desc, k.uo_rt_24hr desc, k.uo_rt_12hr desc, k.uo_rt_6hr desc
+ ) as rn
+ from icustays ie
+ inner join kdigo_stages k
+ on ie.icustay_id = k.icustay_id
+ where (cast(extract(epoch from (k.charttime - ie.intime))/(60*60) as numeric)) > -6
+ and (cast(extract(epoch from (k.charttime - ie.intime))/(60*60*24) as numeric)) <= 7
+ and k.aki_stage_uo is not null
+)
+-- final table is aki_stage, include worst cr/uo for convenience
+select
+ ie.icustay_id
+ , cr.charttime as charttime_creat
+ , cr.creat
+ , cr.aki_stage_creat
+ , uo.charttime as charttime_uo
+ , uo.uo_rt_6hr
+ , uo.uo_rt_12hr
+ , uo.uo_rt_24hr
+ , uo.aki_stage_uo
+
+ -- classify aki using both creatinine/urine output criteria
+ , greatest(
+ coalesce(cr.aki_stage_creat, 0),
+ coalesce(uo.aki_stage_uo, 0)
+ ) as aki_stage_7day
+ , case when cr.aki_stage_creat > 0 or uo.aki_stage_uo > 0 then 1 else 0 end as aki_7day
+
+from icustays ie
+left join cr_aki cr
+ on ie.icustay_id = cr.icustay_id
+ and cr.rn = 1
+left join uo_aki uo
+ on ie.icustay_id = uo.icustay_id
+ and uo.rn = 1
+order by ie.icustay_id;
diff --git a/mimic-iii-carevue/code/concepts/40_kdigo_stages_48hr.sql b/mimic-iii-carevue/code/concepts/40_kdigo_stages_48hr.sql
new file mode 100644
index 000000000..f38286b29
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/40_kdigo_stages_48hr.sql
@@ -0,0 +1,68 @@
+drop table if exists kdigo_stages_48hr; create table kdigo_stages_48hr as
+-- This query checks if the patient had AKI during the first 48 hours of their ICU
+-- stay according to the KDIGO guideline.
+-- https://kdigo.org/wp-content/uploads/2016/10/KDIGO-2012-AKI-Guideline-English.pdf
+
+-- get the worst staging of creatinine in the first 48 hours
+with cr_aki as
+(
+ select
+ k.icustay_id
+ , k.charttime
+ , k.creat
+ , k.aki_stage_creat
+ , row_number() over (partition by k.icustay_id order by k.aki_stage_creat desc, k.creat desc) as rn
+ from icustays ie
+ inner join kdigo_stages k
+ on ie.icustay_id = k.icustay_id
+ where (cast(extract(epoch from (k.charttime - ie.intime))/(60*60) as numeric)) > -6
+ and (cast(extract(epoch from (k.charttime - ie.intime))/(60*60) as numeric)) <= 48
+ and k.aki_stage_creat is not null
+)
+-- get the worst staging of urine output in the first 48 hours
+, uo_aki as
+(
+ select
+ k.icustay_id
+ , k.charttime
+ , k.uo_rt_6hr, k.uo_rt_12hr, k.uo_rt_24hr
+ , k.aki_stage_uo
+ , row_number() over
+ (
+ partition by k.icustay_id
+ order by k.aki_stage_uo desc, k.uo_rt_24hr desc, k.uo_rt_12hr desc, k.uo_rt_6hr desc
+ ) as rn
+ from icustays ie
+ inner join kdigo_stages k
+ on ie.icustay_id = k.icustay_id
+ where (cast(extract(epoch from (k.charttime - ie.intime))/(60*60) as numeric)) > -6
+ and (cast(extract(epoch from (k.charttime - ie.intime))/(60*60) as numeric)) <= 48
+ and k.aki_stage_uo is not null
+)
+-- final table is aki_stage, include worst cr/uo for convenience
+select
+ ie.icustay_id
+ , cr.charttime as charttime_creat
+ , cr.creat
+ , cr.aki_stage_creat
+ , uo.charttime as charttime_uo
+ , uo.uo_rt_6hr
+ , uo.uo_rt_12hr
+ , uo.uo_rt_24hr
+ , uo.aki_stage_uo
+
+ -- classify aki using both creatinine/urine output criteria
+ , greatest(
+ coalesce(cr.aki_stage_creat, 0),
+ coalesce(uo.aki_stage_uo, 0)
+ ) as aki_stage_48hr
+ , case when cr.aki_stage_creat > 0 or uo.aki_stage_uo > 0 then 1 else 0 end as aki_48hr
+
+from icustays ie
+left join cr_aki cr
+ on ie.icustay_id = cr.icustay_id
+ and cr.rn = 1
+left join uo_aki uo
+ on ie.icustay_id = uo.icustay_id
+ and uo.rn = 1
+order by ie.icustay_id;
diff --git a/mimic-iii-carevue/code/concepts/41_meld.sql b/mimic-iii-carevue/code/concepts/41_meld.sql
new file mode 100644
index 000000000..cb0efe8d7
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/41_meld.sql
@@ -0,0 +1,157 @@
+drop table if exists meld; create table meld as
+-- Model for end-stage liver disease (MELD)
+-- This model is used to determine prognosis and receipt of liver transplantation.
+
+-- Reference:
+-- Kamath PS, Wiesner RH, Malinchoc M, Kremers W, Therneau TM,
+-- Kosberg CL, D'Amico G, Dickson ER, Kim WR.
+-- A model to predict survival in patients with end-stage liver disease.
+-- Hepatology. 2001 Feb;33(2):464-70.
+
+
+-- Updated January 2016 to include serum sodium, see:
+-- https://optn.transplant.hrsa.gov/news/meld-serum-sodium-policy-changes/
+
+-- Here is the relevant portion of the policy note:
+-- 9.1.D MELD Score
+-- Candidates who are at least 12 years old receive an initial MELD(i) score equal to:
+-- 0.957 x ln(creatinine mg/dL) + 0.378 x ln(bilirubin mg/dL) + 1.120 x ln(INR) + 0.643
+
+-- Laboratory values less than 1.0 will be set to 1.0 when calculating a candidate’s MELD
+-- score.
+
+-- The following candidates will receive a creatinine value of 4.0 mg/dL:
+-- - Candidates with a creatinine value greater than 4.0 mg/dL
+-- - Candidates who received two or more dialysis treatments within the prior week
+-- - Candidates who received 24 hours of continuous veno-venous hemodialysis (CVVHD) within the prior week
+
+-- The maximum MELD score is 40. The MELD score derived from this calculation will be rounded to the tenth decimal place and then multiplied by 10.
+
+-- For candidates with an initial MELD score greater than 11, The MELD score is then recalculated as follows:
+-- MELD = MELD(i) + 1.32*(137-Na) – [0.033*MELD(i)*(137-Na)]
+-- Sodium values less than 125 mmol/L will be set to 125, and values greater than 137 mmol/L will be set to 137.
+
+
+
+-- TODO needed in this code:
+-- 1. identify 2x dialysis in the past week, or 24 hours of CVVH
+-- at the moment it just checks for any dialysis on the day
+-- 2. adjust the serum sodium using the corresponding glucose measurement
+-- Measured sodium + 0.024 * (Serum glucose - 100) (Hiller, 1999)
+
+with cohort as
+(
+select ie.subject_id, ie.hadm_id, ie.icustay_id
+ , ie.intime
+ , ie.outtime
+
+ , labs.creatinine_max
+ , labs.bilirubin_max
+ , labs.inr_max
+ , labs.sodium_min
+
+ , r.rrt
+
+from icustays ie
+inner join admissions adm
+ on ie.hadm_id = adm.hadm_id
+inner join patients pat
+ on ie.subject_id = pat.subject_id
+
+-- join to custom tables to get more data....
+left join labs_first_day labs
+ on ie.icustay_id = labs.icustay_id
+left join rrt_first_day r
+ on ie.icustay_id = r.icustay_id
+)
+, score as
+(
+ select subject_id, hadm_id, icustay_id
+ , rrt
+ , creatinine_max
+ , bilirubin_max
+ , inr_max
+ , sodium_min
+
+ -- TODO: Corrected Sodium
+ , case
+ when sodium_min is null
+ then 0.0
+ when sodium_min > 137
+ then 0.0
+ when sodium_min < 125
+ then 12.0 -- 137 - 125 = 12
+ else 137.0-sodium_min
+ end as sodium_score
+
+ -- if hemodialysis, value for Creatinine is automatically set to 4.0
+ , case
+ when rrt = 1 or creatinine_max > 4.0
+ then (0.957 * ln(4))
+ -- if creatinine < 1, score is 1
+ when creatinine_max < 1
+ then (0.957 * ln(1))
+ else 0.957 * coalesce(ln(creatinine_max),ln(1))
+ end as creatinine_score
+
+ , case
+ -- if value < 1, score is 1
+ when bilirubin_max < 1
+ then 0.378 * ln(1)
+ else 0.378 * coalesce(ln(bilirubin_max),ln(1))
+ end as bilirubin_score
+
+ , case
+ when inr_max < 1
+ then ( 1.120 * ln(1) + 0.643 )
+ else ( 1.120 * coalesce(ln(inr_max),ln(1)) + 0.643 )
+ end as inr_score
+
+ from cohort
+)
+, score2 as
+(
+ select
+ subject_id, hadm_id, icustay_id
+ , rrt
+ , creatinine_max
+ , bilirubin_max
+ , inr_max
+ , sodium_min
+
+ , creatinine_score
+ , sodium_score
+ , bilirubin_score
+ , inr_score
+
+ , case
+ when (creatinine_score + bilirubin_score + inr_score) > 40
+ then 40.0
+ else
+ round(cast(creatinine_score + bilirubin_score + inr_score as numeric),1)*10
+ end as meld_initial
+ from score
+)
+select
+ subject_id, hadm_id, icustay_id
+
+ -- MELD Score without sodium change
+ , meld_initial
+
+ -- MELD Score (2016) = MELD*10 + 1.32*(137-Na) – [0.033*MELD*10*(137-Na)]
+ , case
+ when meld_initial > 11
+ then meld_initial + 1.32*sodium_score - 0.033*meld_initial*sodium_score
+ else
+ meld_initial
+ end as meld
+
+ -- original variables
+ , rrt
+ , creatinine_max
+ , bilirubin_max
+ , inr_max
+ , sodium_min
+
+from score2
+order by icustay_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/42_oasis.sql b/mimic-iii-carevue/code/concepts/42_oasis.sql
new file mode 100644
index 000000000..04f4c6575
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/42_oasis.sql
@@ -0,0 +1,253 @@
+drop table if exists oasis; create table oasis as
+-- ------------------------------------------------------------------
+-- Title: Oxford Acute Severity of Illness Score (oasis)
+-- This query extracts the Oxford acute severity of illness score.
+-- This score is a measure of severity of illness for patients in the ICU.
+-- The score is calculated on the first day of each ICU patients' stay.
+-- ------------------------------------------------------------------
+
+-- Reference for OASIS:
+-- Johnson, Alistair EW, Andrew A. Kramer, and Gari D. Clifford.
+-- "A new severity of illness scale using a subset of acute physiology and chronic health evaluation data elements shows comparable predictive accuracy*."
+-- Critical care medicine 41, no. 7 (2013): 1711-1718.
+
+-- Variables used in OASIS:
+-- Heart rate, GCS, MAP, Temperature, Respiratory rate, Ventilation status (sourced FROM chartevents)
+-- Urine output (sourced from OUTPUTEVENTS)
+-- Elective surgery (sourced FROM admissions and SERVICES)
+-- Pre-ICU in-hospital length of stay (sourced FROM admissions and ICUSTAYS)
+-- Age (sourced FROM patients)
+
+-- The following views are required to run this query:
+-- 1) urine_output_first_day - generated by urine-output-first-day.sql
+-- 2) vent_first_day - generated by ventilated-first-day.sql
+-- 3) vitals_first_day - generated by vitals-first-day.sql
+-- 4) gcs_first_day - generated by gcs-first-day.sql
+
+
+-- Regarding missing values:
+-- The ventilation flag is always 0/1. It cannot be missing, since VENT=0 if no data is found for vent settings.
+
+-- Note:
+-- The score is calculated for *all* ICU patients, with the assumption that the user will subselect appropriate ICUSTAY_IDs.
+-- For example, the score is calculated for neonates, but it is likely inappropriate to actually use the score values for these patients.
+
+
+with surgflag as
+(
+ select ie.icustay_id
+ , max(case
+ when lower(curr_service) like '%surg%' then 1
+ when curr_service = 'ORTHO' then 1
+ else 0 end) as surgical
+ from icustays ie
+ left join services se
+ on ie.hadm_id = se.hadm_id
+ and se.transfertime < (ie.intime + interval '1 day')
+ group by ie.icustay_id
+)
+, cohort as
+(
+select ie.subject_id, ie.hadm_id, ie.icustay_id
+ , ie.intime
+ , ie.outtime
+ , adm.deathtime
+ , round((cast(extract(epoch from (ie.intime - adm.admittime))/(60) as numeric)),2) as preiculos
+ , round((cast(extract(epoch from (ie.intime - pat.dob))/(60*60*24*365) as numeric)),0) as age
+ , gcs.mingcs
+ , vital.heartrate_max
+ , vital.heartrate_min
+ , vital.meanbp_max
+ , vital.meanbp_min
+ , vital.resprate_max
+ , vital.resprate_min
+ , vital.tempc_max
+ , vital.tempc_min
+ , vent.vent as mechvent
+ , uo.urineoutput
+
+ , case
+ when adm.admission_type = 'ELECTIVE' and sf.surgical = 1
+ then 1
+ when adm.admission_type is null or sf.surgical is null
+ then null
+ else 0
+ end as electivesurgery
+
+ -- age group
+ , case
+ when (cast(extract(epoch from (ie.intime - pat.dob))/(60*60*24*365) as numeric)) <= 1 then 'neonate'
+ when (cast(extract(epoch from (ie.intime - pat.dob))/(60*60*24*365) as numeric)) <= 15 then 'middle'
+ else 'adult' end as icustay_age_group
+
+ -- mortality flags
+ , case
+ when adm.deathtime between ie.intime and ie.outtime
+ then 1
+ when adm.deathtime <= ie.intime -- sometimes there are typographical errors in the death date
+ then 1
+ when adm.dischtime <= ie.outtime and adm.discharge_location = 'DEAD/EXPIRED'
+ then 1
+ else 0 end
+ as icustay_expire_flag
+ , adm.hospital_expire_flag
+from icustays ie
+inner join admissions adm
+ on ie.hadm_id = adm.hadm_id
+inner join patients pat
+ on ie.subject_id = pat.subject_id
+left join surgflag sf
+ on ie.icustay_id = sf.icustay_id
+-- join to custom tables to get more data....
+left join gcs_first_day gcs
+ on ie.icustay_id = gcs.icustay_id
+left join vitals_first_day vital
+ on ie.icustay_id = vital.icustay_id
+left join urine_output_first_day uo
+ on ie.icustay_id = uo.icustay_id
+left join ventilation_first_day vent
+ on ie.icustay_id = vent.icustay_id
+)
+, scorecomp as
+(
+select co.subject_id, co.hadm_id, co.icustay_id
+, co.icustay_age_group
+, co.icustay_expire_flag
+, co.hospital_expire_flag
+
+-- Below code calculates the component scores needed for oasis
+, case when preiculos is null then null
+ when preiculos < 10.2 then 5
+ when preiculos < 297 then 3
+ when preiculos < 1440 then 0
+ when preiculos < 18708 then 1
+ else 2 end as preiculos_score
+, case when age is null then null
+ when age < 24 then 0
+ when age <= 53 then 3
+ when age <= 77 then 6
+ when age <= 89 then 9
+ when age >= 90 then 7
+ else 0 end as age_score
+, case when mingcs is null then null
+ when mingcs <= 7 then 10
+ when mingcs < 14 then 4
+ when mingcs = 14 then 3
+ else 0 end as gcs_score
+, case when heartrate_max is null then null
+ when heartrate_max > 125 then 6
+ when heartrate_min < 33 then 4
+ when heartrate_max >= 107 and heartrate_max <= 125 then 3
+ when heartrate_max >= 89 and heartrate_max <= 106 then 1
+ else 0 end as heartrate_score
+, case when meanbp_min is null then null
+ when meanbp_min < 20.65 then 4
+ when meanbp_min < 51 then 3
+ when meanbp_max > 143.44 then 3
+ when meanbp_min >= 51 and meanbp_min < 61.33 then 2
+ else 0 end as meanbp_score
+, case when resprate_min is null then null
+ when resprate_min < 6 then 10
+ when resprate_max > 44 then 9
+ when resprate_max > 30 then 6
+ when resprate_max > 22 then 1
+ when resprate_min < 13 then 1 else 0
+ end as resprate_score
+, case when tempc_max is null then null
+ when tempc_max > 39.88 then 6
+ when tempc_min >= 33.22 and tempc_min <= 35.93 then 4
+ when tempc_max >= 33.22 and tempc_max <= 35.93 then 4
+ when tempc_min < 33.22 then 3
+ when tempc_min > 35.93 and tempc_min <= 36.39 then 2
+ when tempc_max >= 36.89 and tempc_max <= 39.88 then 2
+ else 0 end as temp_score
+, case when UrineOutput is null then null
+ when UrineOutput < 671.09 then 10
+ when UrineOutput > 6896.80 then 8
+ when UrineOutput >= 671.09
+ and UrineOutput <= 1426.99 then 5
+ when UrineOutput >= 1427.00
+ and UrineOutput <= 2544.14 then 1
+ else 0 end as urineoutput_score
+, case when mechvent is null then null
+ when mechvent = 1 then 9
+ else 0 end as mechvent_score
+, case when electivesurgery is null then null
+ when electivesurgery = 1 then 0
+ else 6 end as electivesurgery_score
+
+
+-- The below code gives the component associated with each score
+-- This is not needed to calculate oasis, but provided for user convenience.
+-- If both the min/max are in the normal range (score of 0), then the average value is stored.
+, preiculos
+, age
+, mingcs as gcs
+, case when heartrate_max is null then null
+ when heartrate_max > 125 then heartrate_max
+ when heartrate_min < 33 then heartrate_min
+ when heartrate_max >= 107 and heartrate_max <= 125 then heartrate_max
+ when heartrate_max >= 89 and heartrate_max <= 106 then heartrate_max
+ else (heartrate_min+heartrate_max)/2 end as heartrate
+, case when meanbp_min is null then null
+ when meanbp_min < 20.65 then meanbp_min
+ when meanbp_min < 51 then meanbp_min
+ when meanbp_max > 143.44 then meanbp_max
+ when meanbp_min >= 51 and meanbp_min < 61.33 then meanbp_min
+ else (meanbp_min+meanbp_max)/2 end as meanbp
+, case when resprate_min is null then null
+ when resprate_min < 6 then resprate_min
+ when resprate_max > 44 then resprate_max
+ when resprate_max > 30 then resprate_max
+ when resprate_max > 22 then resprate_max
+ when resprate_min < 13 then resprate_min
+ else (resprate_min+resprate_max)/2 end as resprate
+, case when tempc_max is null then null
+ when tempc_max > 39.88 then tempc_max
+ when tempc_min >= 33.22 and tempc_min <= 35.93 then tempc_min
+ when tempc_max >= 33.22 and tempc_max <= 35.93 then tempc_max
+ when tempc_min < 33.22 then tempc_min
+ when tempc_min > 35.93 and tempc_min <= 36.39 then tempc_min
+ when tempc_max >= 36.89 and tempc_max <= 39.88 then tempc_max
+ else (tempc_min+tempc_max)/2 end as temp
+, urineoutput
+, mechvent
+, electivesurgery
+from cohort co
+)
+, score as
+(
+select s.*
+ , coalesce(age_score,0)
+ + coalesce(preiculos_score,0)
+ + coalesce(gcs_score,0)
+ + coalesce(heartrate_score,0)
+ + coalesce(meanbp_score,0)
+ + coalesce(resprate_score,0)
+ + coalesce(temp_score,0)
+ + coalesce(urineoutput_score,0)
+ + coalesce(mechvent_score,0)
+ + coalesce(electivesurgery_score,0)
+ as oasis
+from scorecomp s
+)
+select
+ subject_id, hadm_id, icustay_id
+ , icustay_age_group
+ , hospital_expire_flag
+ , icustay_expire_flag
+ , oasis
+ -- Calculate the probability of in-hospital mortality
+ , 1 / (1 + exp(- (-6.1746 + 0.1275*(oasis) ))) as oasis_prob
+ , age, age_score
+ , preiculos, preiculos_score
+ , gcs, gcs_score
+ , heartrate, heartrate_score
+ , meanbp, meanbp_score
+ , resprate, resprate_score
+ , temp, temp_score
+ , urineoutput, urineoutput_score
+ , mechvent, mechvent_score
+ , electivesurgery, electivesurgery_score
+from score
+order by icustay_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/43_sofa.sql b/mimic-iii-carevue/code/concepts/43_sofa.sql
new file mode 100644
index 000000000..f14bd8ae7
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/43_sofa.sql
@@ -0,0 +1,242 @@
+drop table if exists sofa; create table sofa as
+-- ------------------------------------------------------------------
+-- Title: Sequential Organ Failure Assessment (SOFA)
+-- This query extracts the sequential organ failure assessment (formally: sepsis-related organ failure assessment).
+-- This score is a measure of organ failure for patients in the ICU.
+-- The score is calculated on the first day of each ICU patients' stay.
+-- ------------------------------------------------------------------
+
+-- Reference for SOFA:
+-- Jean-Louis Vincent, Rui Moreno, Jukka Takala, Sheila Willatts, Arnaldo De Mendonça,
+-- Hajo Bruining, C. K. Reinhart, Peter M Suter, and L. G. Thijs.
+-- "The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure."
+-- Intensive care medicine 22, no. 7 (1996): 707-710.
+
+-- Variables used in SOFA:
+-- GCS, MAP, FiO2, Ventilation status (sourced FROM chartevents)
+-- Creatinine, Bilirubin, FiO2, PaO2, Platelets (sourced FROM labevents)
+-- Dobutamine, Epinephrine, Norepinephrine (sourced FROM inputevents_mv and INPUTEVENTS_CV)
+-- Urine output (sourced from OUTPUTEVENTS)
+
+-- The following views required to run this query:
+-- 1) urine_output_first_day - generated by urine-output-first-day.sql
+-- 2) vitals_first_day - generated by vitals-first-day.sql
+-- 3) gcs_first_day - generated by gcs-first-day.sql
+-- 4) labs_first_day - generated by labs-first-day.sql
+-- 5) blood_gas_first_day_arterial - generated by blood-gas-first-day-arterial.sql
+-- 6) ventilation_durations - generated by ventilation_durations.sql
+
+-- Note:
+-- The score is calculated for *all* ICU patients, with the assumption that the user will subselect appropriate ICUSTAY_IDs.
+-- For example, the score is calculated for neonates, but it is likely inappropriate to actually use the score values for these patients.
+
+with wt as
+(
+ select ie.icustay_id
+ -- ensure weight is measured in kg
+ , avg(case
+ when itemid in (762, 763, 3723, 3580, 226512)
+ then valuenum
+ -- convert lbs to kgs
+ when itemid in (3581)
+ then valuenum * 0.45359237
+ when itemid in (3582)
+ then valuenum * 0.0283495231
+ else null
+ end) as weight
+
+ from icustays ie
+ left join chartevents c
+ on ie.icustay_id = c.icustay_id
+ where valuenum is not null
+ and itemid in
+ (
+ 762, 763, 3723, 3580, -- Weight Kg
+ 3581, -- Weight lb
+ 3582, -- Weight oz
+ 226512 -- Metavision: Admission Weight (Kg)
+ )
+ and valuenum != 0
+ and charttime between (ie.intime - interval '1 day') and (ie.intime + interval '1 day')
+ -- exclude rows marked as error
+ and (c.error is null or c.error = 0)
+ group by ie.icustay_id
+)
+
+, vaso_cv as
+(
+ select ie.icustay_id
+ -- case statement determining whether the ITEMID is an instance of vasopressor usage
+ , max(case
+ when itemid = 30047 then rate / wt.weight -- measured in mcgmin
+ when itemid = 30120 then rate -- measured in mcgkgmin ** there are clear errors, perhaps actually mcgmin
+ else null
+ end) as rate_norepinephrine
+
+ , max(case
+ when itemid = 30044 then rate / wt.weight -- measured in mcgmin
+ when itemid in (30119,30309) then rate -- measured in mcgkgmin
+ else null
+ end) as rate_epinephrine
+
+ , max(case when itemid in (30043,30307) then rate end) as rate_dopamine
+ , max(case when itemid in (30042,30306) then rate end) as rate_dobutamine
+
+ from icustays ie
+ inner join inputevents_cv cv
+ on ie.icustay_id = cv.icustay_id and cv.charttime between ie.intime and (ie.intime + interval '1 day')
+ left join wt
+ on ie.icustay_id = wt.icustay_id
+ where itemid in (30047,30120,30044,30119,30309,30043,30307,30042,30306)
+ and rate is not null
+ group by ie.icustay_id
+)
+
+, pafi1 as
+(
+ -- join blood gas to ventilation durations to determine if patient was vent
+ select bg.icustay_id, bg.charttime
+ , pao2fio2
+ , case when vd.icustay_id is not null then 1 else 0 end as isvent
+ from blood_gas_first_day_arterial bg
+ left join ventilation_durations vd
+ on bg.icustay_id = vd.icustay_id
+ and bg.charttime >= vd.starttime
+ and bg.charttime <= vd.endtime
+ order by bg.icustay_id, bg.charttime
+)
+, pafi2 as
+(
+ -- because pafi has an interaction between vent/PaO2:FiO2, we need two columns for the score
+ -- it can happen that the lowest unventilated PaO2/FiO2 is 68, but the lowest ventilated PaO2/FiO2 is 120
+ -- in this case, the SOFA score is 3, *not* 4.
+ select icustay_id
+ , min(case when isvent = 0 then pao2fio2 else null end) as pao2fio2_novent_min
+ , min(case when isvent = 1 then pao2fio2 else null end) as pao2fio2_vent_min
+ from pafi1
+ group by icustay_id
+)
+-- Aggregate the components for the score
+, scorecomp as
+(
+select ie.icustay_id
+ , v.meanbp_min
+ , cv.rate_norepinephrine
+ , cv.rate_epinephrine
+ , cv.rate_dopamine
+ , cv.rate_dobutamine
+
+ , l.creatinine_max
+ , l.bilirubin_max
+ , l.platelet_min
+
+ , pf.pao2fio2_novent_min
+ , pf.pao2fio2_vent_min
+
+ , uo.urineoutput
+
+ , gcs.mingcs
+from icustays ie
+left join vaso_cv cv
+ on ie.icustay_id = cv.icustay_id
+left join pafi2 pf
+ on ie.icustay_id = pf.icustay_id
+left join vitals_first_day v
+ on ie.icustay_id = v.icustay_id
+left join labs_first_day l
+ on ie.icustay_id = l.icustay_id
+left join urine_output_first_day uo
+ on ie.icustay_id = uo.icustay_id
+left join gcs_first_day gcs
+ on ie.icustay_id = gcs.icustay_id
+)
+, scorecalc as
+(
+ -- Calculate the final score
+ -- note that if the underlying data is missing, the component is null
+ -- eventually these are treated as 0 (normal), but knowing when data is missing is useful for debugging
+ select icustay_id
+ -- Respiration
+ , case
+ when pao2fio2_vent_min < 100 then 4
+ when pao2fio2_vent_min < 200 then 3
+ when pao2fio2_novent_min < 300 then 2
+ when pao2fio2_novent_min < 400 then 1
+ when coalesce(pao2fio2_vent_min, pao2fio2_novent_min) is null then null
+ else 0
+ end as respiration
+
+ -- Coagulation
+ , case
+ when platelet_min < 20 then 4
+ when platelet_min < 50 then 3
+ when platelet_min < 100 then 2
+ when platelet_min < 150 then 1
+ when platelet_min is null then null
+ else 0
+ end as coagulation
+
+ -- Liver
+ , case
+ -- Bilirubin checks in mg/dL
+ when bilirubin_max >= 12.0 then 4
+ when bilirubin_max >= 6.0 then 3
+ when bilirubin_max >= 2.0 then 2
+ when bilirubin_max >= 1.2 then 1
+ when bilirubin_max is null then null
+ else 0
+ end as liver
+
+ -- Cardiovascular
+ , case
+ when rate_dopamine > 15 or rate_epinephrine > 0.1 or rate_norepinephrine > 0.1 then 4
+ when rate_dopamine > 5 or rate_epinephrine <= 0.1 or rate_norepinephrine <= 0.1 then 3
+ when rate_dopamine > 0 or rate_dobutamine > 0 then 2
+ when meanbp_min < 70 then 1
+ when coalesce(meanbp_min, rate_dopamine, rate_dobutamine, rate_epinephrine, rate_norepinephrine) is null then null
+ else 0
+ end as cardiovascular
+
+ -- Neurological failure (GCS)
+ , case
+ when (mingcs >= 13 and mingcs <= 14) then 1
+ when (mingcs >= 10 and mingcs <= 12) then 2
+ when (mingcs >= 6 and mingcs <= 9) then 3
+ when mingcs < 6 then 4
+ when mingcs is null then null
+ else 0 end
+ as cns
+
+ -- Renal failure - high creatinine or low urine output
+ , case
+ when (creatinine_max >= 5.0) then 4
+ when urineoutput < 200 then 4
+ when (creatinine_max >= 3.5 and creatinine_max < 5.0) then 3
+ when urineoutput < 500 then 3
+ when (creatinine_max >= 2.0 and creatinine_max < 3.5) then 2
+ when (creatinine_max >= 1.2 and creatinine_max < 2.0) then 1
+ when coalesce(urineoutput, creatinine_max) is null then null
+ else 0 end
+ as renal
+ from scorecomp
+)
+select ie.subject_id, ie.hadm_id, ie.icustay_id
+ -- Combine all the scores to get SOFA
+ -- Impute 0 if the score is missing
+ , coalesce(respiration,0)
+ + coalesce(coagulation,0)
+ + coalesce(liver,0)
+ + coalesce(cardiovascular,0)
+ + coalesce(cns,0)
+ + coalesce(renal,0)
+ as sofa
+, respiration
+, coagulation
+, liver
+, cardiovascular
+, cns
+, renal
+from icustays ie
+left join scorecalc s
+ on ie.icustay_id = s.icustay_id
+order by ie.icustay_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/44_saps.sql b/mimic-iii-carevue/code/concepts/44_saps.sql
new file mode 100644
index 000000000..ce794ea12
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/44_saps.sql
@@ -0,0 +1,335 @@
+drop table if exists saps; create table saps as
+-- ------------------------------------------------------------------
+-- Title: Simplified Acute Physiology Score (SAPS)
+-- This query extracts the simplified acute physiology score.
+-- This score is a measure of patient severity of illness.
+-- The score is calculated on the first day of each ICU patients' stay.
+-- ------------------------------------------------------------------
+
+-- Reference for SAPS:
+-- Jean-Roger Le Gall, Philippe Loirat, Annick Alperovitch, Paul Glaser, Claude Granthil,
+-- Daniel Mathieu, Philippe Mercier, Remi Thomas, and Daniel Villers.
+-- "A simplified acute physiology score for ICU patients."
+-- Critical care medicine 12, no. 11 (1984): 975-977.
+
+-- Variables used in SAPS:
+-- Age, GCS
+-- VITALS: Heart rate, systolic blood pressure, temperature, respiration rate
+-- FLAGS: ventilation/cpap
+-- IO: urine output
+-- LABS: blood urea nitrogen, hematocrit, WBC, glucose, potassium, sodium, HCO3
+
+-- The following views are required to run this query:
+-- 1) urine_output_first_day - generated by urine-output-first-day.sql
+-- 2) vent_first_day - generated by ventilated-first-day.sql
+-- 3) vitals_first_day - generated by vitals-first-day.sql
+-- 4) gcs_first_day - generated by gcs-first-day.sql
+-- 5) labs_first_day - generated by labs-first-day.sql
+
+-- Note:
+-- The score is calculated for *all* ICU patients, with the assumption that the user will subselect appropriate icustay_ids.
+-- For example, the score is calculated for neonates, but it is likely inappropriate to actually use the score values for these patients.
+
+-- extract CPAP from the "Oxygen Delivery Device" fields
+with cpap as
+(
+ select ie.icustay_id
+ , max(case
+ when lower(ce.value) like '%cpap%' then 1
+ when lower(ce.value) like '%bipap mask%' then 1
+ else 0 end) as cpap
+ from icustays ie
+ inner join chartevents ce
+ on ie.icustay_id = ce.icustay_id
+ and ce.charttime between ie.intime and (ie.intime + interval '1 day')
+ where itemid in
+ (
+ -- TODO: when metavision data import fixed, check the values in 226732 match the value clause below
+ 467, 469, 226732
+ )
+ and (lower(ce.value) like '%cpap%' or lower(ce.value) like '%bipap mask%')
+ -- exclude rows marked as error
+ AND (ce.error IS NULL OR ce.error = 0)
+ group by ie.icustay_id
+)
+, cohort as
+(
+select ie.subject_id, ie.hadm_id, ie.icustay_id
+ , ie.intime
+ , ie.outtime
+
+ -- the casts ensure the result is numeric.. we could equally extract EPOCH from the interval
+ -- however this code works in Oracle and Postgres
+ , round((cast(extract(epoch from (ie.intime - pat.dob))/(60*60*24*365) as numeric)),0) as age
+ , gcs.mingcs
+ , vital.heartrate_max
+ , vital.heartrate_min
+ , vital.sysbp_max
+ , vital.sysbp_min
+ , vital.resprate_max
+ , vital.resprate_min
+ , vital.tempc_max
+ , vital.tempc_min
+
+ , coalesce(vital.glucose_max, labs.glucose_max) as glucose_max
+ , coalesce(vital.glucose_min, labs.glucose_min) as glucose_min
+
+ , labs.bun_max
+ , labs.bun_min
+ , labs.hematocrit_max
+ , labs.hematocrit_min
+ , labs.wbc_max
+ , labs.wbc_min
+ , labs.sodium_max
+ , labs.sodium_min
+ , labs.potassium_max
+ , labs.potassium_min
+ , labs.bicarbonate_max
+ , labs.bicarbonate_min
+
+ , vent.vent as mechvent
+ , uo.urineoutput
+
+ , cp.cpap
+
+from icustays ie
+inner join admissions adm
+ on ie.hadm_id = adm.hadm_id
+inner join patients pat
+ on ie.subject_id = pat.subject_id
+
+-- join to above view to get CPAP
+left join cpap cp
+ on ie.icustay_id = cp.icustay_id
+
+-- join to custom tables to get more data....
+left join gcs_first_day gcs
+ on ie.icustay_id = gcs.icustay_id
+left join vitals_first_day vital
+ on ie.icustay_id = vital.icustay_id
+left join urine_output_first_day uo
+ on ie.icustay_id = uo.icustay_id
+left join ventilation_first_day vent
+ on ie.icustay_id = vent.icustay_id
+left join labs_first_day labs
+ on ie.icustay_id = labs.icustay_id
+)
+, scorecomp as
+(
+select
+ cohort.*
+ -- Below code calculates the component scores needed for SAPS
+ , case
+ when age is null then null
+ when age <= 45 then 0
+ when age <= 55 then 1
+ when age <= 65 then 2
+ when age <= 75 then 3
+ when age > 75 then 4
+ end as age_score
+ , case
+ when heartrate_max is null then null
+ when heartrate_max >= 180 then 4
+ when heartrate_min < 40 then 4
+ when heartrate_max >= 140 then 3
+ when heartrate_min <= 54 then 3
+ when heartrate_max >= 110 then 2
+ when heartrate_min <= 69 then 2
+ when heartrate_max >= 70 and heartrate_max <= 109
+ and heartrate_min >= 70 and heartrate_min <= 109
+ then 0
+ end as hr_score
+ , case
+ when sysbp_min is null then null
+ when sysbp_max >= 190 then 4
+ when sysbp_min < 55 then 4
+ when sysbp_max >= 150 then 2
+ when sysbp_min <= 79 then 2
+ when sysbp_max >= 80 and sysbp_max <= 149
+ and sysbp_min >= 80 and sysbp_min <= 149
+ then 0
+ end as sysbp_score
+
+ , case
+ when tempc_max is null then null
+ when tempc_max >= 41.0 then 4
+ when tempc_min < 30.0 then 4
+ when tempc_max >= 39.0 then 3
+ when tempc_min <= 31.9 then 3
+ when tempc_min <= 33.9 then 2
+ when tempc_max > 38.4 then 1
+ when tempc_min < 36.0 then 1
+ when tempc_max >= 36.0 and tempc_max <= 38.4
+ and tempc_min >= 36.0 and tempc_min <= 38.4
+ then 0
+ end as temp_score
+
+ , case
+ when resprate_min is null then null
+ when resprate_max >= 50 then 4
+ when resprate_min < 6 then 4
+ when resprate_max >= 35 then 3
+ when resprate_min <= 9 then 2
+ when resprate_max >= 25 then 1
+ when resprate_min <= 11 then 1
+ when resprate_max >= 12 and resprate_max <= 24
+ and resprate_min >= 12 and resprate_min <= 24
+ then 0
+ end as resp_score
+
+ , case
+ when coalesce(mechvent,cpap) is null then null
+ when cpap = 1 then 3
+ when mechvent = 1 then 3
+ else 0
+ end as vent_score
+
+ , case
+ when urineoutput is null then null
+ when urineoutput > 5000.0 then 2
+ when urineoutput >= 3500.0 then 1
+ when urineoutput >= 700.0 then 0
+ when urineoutput >= 500.0 then 2
+ when urineoutput >= 200.0 then 3
+ when urineoutput < 200.0 then 4
+ end as uo_score
+
+ , case
+ when bun_max is null then null
+ when bun_max >= 55.0 then 4
+ when bun_max >= 36.0 then 3
+ when bun_max >= 29.0 then 2
+ when bun_max >= 7.50 then 1
+ when bun_min < 3.5 then 1
+ when bun_max >= 3.5 and bun_max < 7.5
+ and bun_min >= 3.5 and bun_min < 7.5
+ then 0
+ end as bun_score
+
+ , case
+ when hematocrit_max is null then null
+ when hematocrit_max >= 60.0 then 4
+ when hematocrit_min < 20.0 then 4
+ when hematocrit_max >= 50.0 then 2
+ when hematocrit_min < 30.0 then 2
+ when hematocrit_max >= 46.0 then 1
+ when hematocrit_max >= 30.0 and hematocrit_max < 46.0
+ and hematocrit_min >= 30.0 and hematocrit_min < 46.0
+ then 0
+ end as hematocrit_score
+
+ , case
+ when wbc_max is null then null
+ when wbc_max >= 40.0 then 4
+ when wbc_min < 1.0 then 4
+ when wbc_max >= 20.0 then 2
+ when wbc_min < 3.0 then 2
+ when wbc_max >= 15.0 then 1
+ when wbc_max >= 3.0 and wbc_max < 15.0
+ and wbc_min >= 3.0 and wbc_min < 15.0
+ then 0
+ end as wbc_score
+
+ , case
+ when glucose_max is null then null
+ when glucose_max >= 44.5 then 4
+ when glucose_min < 1.6 then 4
+ when glucose_max >= 27.8 then 3
+ when glucose_min < 2.8 then 3
+ when glucose_min < 3.9 then 2
+ when glucose_max >= 14.0 then 1
+ when glucose_max >= 3.9 and glucose_max < 14.0
+ and glucose_min >= 3.9 and glucose_min < 14.0
+ then 0
+ end as glucose_score
+
+ , case
+ when potassium_max is null then null
+ when potassium_max >= 7.0 then 4
+ when potassium_min < 2.5 then 4
+ when potassium_max >= 6.0 then 3
+ when potassium_min < 3.0 then 2
+ when potassium_max >= 5.5 then 1
+ when potassium_min < 3.5 then 1
+ when potassium_max >= 3.5 and potassium_max < 5.5
+ and potassium_min >= 3.5 and potassium_min < 5.5
+ then 0
+ end as potassium_score
+
+ , case
+ when sodium_max is null then null
+ when sodium_max >= 180 then 4
+ when sodium_min < 110 then 4
+ when sodium_max >= 161 then 3
+ when sodium_min < 120 then 3
+ when sodium_max >= 156 then 2
+ when sodium_min < 130 then 2
+ when sodium_max >= 151 then 1
+ when sodium_max >= 130 and sodium_max < 151
+ and sodium_min >= 130 and sodium_min < 151
+ then 0
+ end as sodium_score
+
+ , case
+ when bicarbonate_max is null then null
+ when bicarbonate_min < 5.0 then 4
+ when bicarbonate_max >= 40.0 then 3
+ when bicarbonate_min < 10.0 then 3
+ when bicarbonate_max >= 30.0 then 1
+ when bicarbonate_min < 20.0 then 1
+ when bicarbonate_max >= 20.0 and bicarbonate_max < 30.0
+ and bicarbonate_min >= 20.0 and bicarbonate_min < 30.0
+ then 0
+ end as bicarbonate_score
+
+ , case
+ when mingcs is null then null
+ when mingcs < 3 then null -- erroneous value/on trach
+ when mingcs = 3 then 4
+ when mingcs < 7 then 3
+ when mingcs < 10 then 2
+ when mingcs < 13 then 1
+ when mingcs >= 13
+ and mingcs <= 15
+ then 0
+ end as gcs_score
+from cohort
+)
+select ie.subject_id, ie.hadm_id, ie.icustay_id
+-- coalesce statements impute normal score of zero if data element is missing
+, coalesce(age_score,0)
++ coalesce(hr_score,0)
++ coalesce(sysbp_score,0)
++ coalesce(resp_score,0)
++ coalesce(temp_score,0)
++ coalesce(uo_score,0)
++ coalesce(vent_score,0)
++ coalesce(bun_score,0)
++ coalesce(hematocrit_score,0)
++ coalesce(wbc_score,0)
++ coalesce(glucose_score,0)
++ coalesce(potassium_score,0)
++ coalesce(sodium_score,0)
++ coalesce(bicarbonate_score,0)
++ coalesce(gcs_score,0)
+ as saps
+, age_score
+, hr_score
+, sysbp_score
+, resp_score
+, temp_score
+, uo_score
+, vent_score
+, bun_score
+, hematocrit_score
+, wbc_score
+, glucose_score
+, potassium_score
+, sodium_score
+, bicarbonate_score
+, gcs_score
+
+from icustays ie
+left join scorecomp s
+ on ie.icustay_id = s.icustay_id
+order by ie.icustay_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/45_sapsii.sql b/mimic-iii-carevue/code/concepts/45_sapsii.sql
new file mode 100644
index 000000000..7cbd6043e
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/45_sapsii.sql
@@ -0,0 +1,383 @@
+drop table if exists sapsii; create table sapsii as
+-- ------------------------------------------------------------------
+-- Title: Simplified Acute Physiology Score II (SAPS II)
+-- This query extracts the simplified acute physiology score II.
+-- This score is a measure of patient severity of illness.
+-- The score is calculated on the first day of each ICU patients' stay.
+-- ------------------------------------------------------------------
+
+-- Reference for SAPS II:
+-- Le Gall, Jean-Roger, Stanley Lemeshow, and Fabienne Saulnier.
+-- "A new simplified acute physiology score (SAPS II) based on a European/North American multicenter study."
+-- JAMA 270, no. 24 (1993): 2957-2963.
+
+-- Variables used in SAPS II:
+-- Age, GCS
+-- VITALS: Heart rate, systolic blood pressure, temperature
+-- FLAGS: ventilation/cpap
+-- IO: urine output
+-- LABS: PaO2/FiO2 ratio, blood urea nitrogen, WBC, potassium, sodium, HCO3
+
+-- The following views are required to run this query:
+-- 1) urine_output_first_day - generated by urine-output-first-day.sql
+-- 2) ventilation_durations - generated by ventilation_durations.sql
+-- 3) vitals_first_day - generated by vitals-first-day.sql
+-- 4) gcs_first_day - generated by gcs-first-day.sql
+-- 5) labs_first_day - generated by labs-first-day.sql
+-- 6) blood_gas_arterial_first_day - generated by blood-gas-first-day-arterial.sql
+
+-- Note:
+-- The score is calculated for *all* ICU patients, with the assumption that the user will subselect appropriate ICUSTAY_IDs.
+-- For example, the score is calculated for neonates, but it is likely inappropriate to actually use the score values for these patients.
+
+-- extract CPAP from the "Oxygen Delivery Device" fields
+with cpap as
+(
+ select ie.icustay_id
+ , min(charttime - interval '1 hour') as starttime
+ , max(charttime + interval '4 hour') as endtime
+ , max(case
+ when lower(ce.value) like '%cpap%' then 1
+ when lower(ce.value) like '%bipap mask%' then 1
+ else 0 end) as cpap
+ from icustays ie
+ inner join chartevents ce
+ on ie.icustay_id = ce.icustay_id
+ and ce.charttime between ie.intime and (ie.intime + interval '1 day')
+ where itemid in
+ (
+ -- TODO: when metavision data import fixed, check the values in 226732 match the value clause below
+ 467, 469, 226732
+ )
+ and (lower(ce.value) like '%cpap%' or lower(ce.value) like '%bipap mask%')
+ -- exclude rows marked as error
+ and (ce.error is null or ce.error = 0)
+ group by ie.icustay_id
+)
+-- extract a flag for surgical service
+-- this combined with "elective" FROM admissions table defines elective/non-elective surgery
+, surgflag as
+(
+ select adm.hadm_id
+ , case when lower(curr_service) like '%surg%' then 1 else 0 end as surgical
+ , row_number() over
+ (
+ partition by adm.hadm_id
+ order by transfertime
+ ) as serviceorder
+ from admissions adm
+ left join services se
+ on adm.hadm_id = se.hadm_id
+)
+-- icd-9 diagnostic codes are our best source for comorbidity information
+-- unfortunately, they are technically a-causal
+-- however, this shouldn't matter too much for the SAPS II comorbidities
+, comorb as
+(
+select hadm_id
+-- these are slightly different than elixhauser comorbidities, but based on them
+-- they include some non-comorbid ICD-9 codes (e.g. 20302, relapse of multiple myeloma)
+ , max(case
+ when substr(icd9_code,1,3) between '042' and '044' then 1
+ end) as aids /* HIV and AIDS */
+ , max(case
+ when icd9_code between '20000' and '20238' then 1 -- lymphoma
+ when icd9_code between '20240' and '20248' then 1 -- leukemia
+ when icd9_code between '20250' and '20302' then 1 -- lymphoma
+ when icd9_code between '20310' and '20312' then 1 -- leukemia
+ when icd9_code between '20302' and '20382' then 1 -- lymphoma
+ when icd9_code between '20400' and '20522' then 1 -- chronic leukemia
+ when icd9_code between '20580' and '20702' then 1 -- other myeloid leukemia
+ when icd9_code between '20720' and '20892' then 1 -- other myeloid leukemia
+ when substr(icd9_code,1,4) = '2386' then 1 -- lymphoma
+ when substr(icd9_code,1,4) = '2733' then 1 -- lymphoma
+ end) as hem
+ , max(case
+ when substr(icd9_code,1,4) between '1960' and '1991' then 1
+ when icd9_code between '20970' and '20975' then 1
+ when icd9_code = '20979' then 1
+ when icd9_code = '78951' then 1
+ end) as mets /* Metastatic cancer */
+ from diagnoses_icd
+ group by hadm_id
+)
+, pafi1 as
+(
+ -- join blood gas to ventilation durations to determine if patient was vent
+ -- also join to cpap table for the same purpose
+ select bg.icustay_id, bg.charttime
+ , pao2fio2
+ , case when vd.icustay_id is not null then 1 else 0 end as vent
+ , case when cp.icustay_id is not null then 1 else 0 end as cpap
+ from blood_gas_first_day_arterial bg
+ left join ventilation_durations vd
+ on bg.icustay_id = vd.icustay_id
+ and bg.charttime >= vd.starttime
+ and bg.charttime <= vd.endtime
+ left join cpap cp
+ on bg.icustay_id = cp.icustay_id
+ and bg.charttime >= cp.starttime
+ and bg.charttime <= cp.endtime
+)
+, pafi2 as
+(
+ -- get the minimum PaO2/FiO2 ratio *only for ventilated/cpap patients*
+ select icustay_id
+ , min(pao2fio2) as pao2fio2_vent_min
+ from pafi1
+ where vent = 1 or cpap = 1
+ group by icustay_id
+)
+, cohort as
+(
+select ie.subject_id, ie.hadm_id, ie.icustay_id
+ , ie.intime
+ , ie.outtime
+
+ -- the casts ensure the result is numeric.. we could equally extract EPOCH from the interval
+ -- however this code works in Oracle and Postgres
+ , round((cast(extract(epoch from (ie.intime - pat.dob))/(60*60*24*365) as numeric)),0) as age
+
+ , vital.heartrate_max
+ , vital.heartrate_min
+ , vital.sysbp_max
+ , vital.sysbp_min
+ , vital.tempc_max
+ , vital.tempc_min
+
+ -- this value is non-null iff the patient is on vent/cpap
+ , pf.pao2fio2_vent_min
+
+ , uo.urineoutput
+
+ , labs.bun_min
+ , labs.bun_max
+ , labs.wbc_min
+ , labs.wbc_max
+ , labs.potassium_min
+ , labs.potassium_max
+ , labs.sodium_min
+ , labs.sodium_max
+ , labs.bicarbonate_min
+ , labs.bicarbonate_max
+ , labs.bilirubin_min
+ , labs.bilirubin_max
+
+ , gcs.mingcs
+
+ , comorb.aids
+ , comorb.hem
+ , comorb.mets
+
+ , case
+ when adm.admission_type = 'ELECTIVE' and sf.surgical = 1
+ then 'ScheduledSurgical'
+ when adm.admission_type != 'ELECTIVE' and sf.surgical = 1
+ then 'UnscheduledSurgical'
+ else 'Medical'
+ end as admissiontype
+
+
+from icustays ie
+inner join admissions adm
+ on ie.hadm_id = adm.hadm_id
+inner join patients pat
+ on ie.subject_id = pat.subject_id
+
+-- join to above views
+left join pafi2 pf
+ on ie.icustay_id = pf.icustay_id
+left join surgflag sf
+ on adm.hadm_id = sf.hadm_id and sf.serviceorder = 1
+left join comorb
+ on ie.hadm_id = comorb.hadm_id
+
+-- join to custom tables to get more data....
+left join gcs_first_day gcs
+ on ie.icustay_id = gcs.icustay_id
+left join vitals_first_day vital
+ on ie.icustay_id = vital.icustay_id
+left join urine_output_first_day uo
+ on ie.icustay_id = uo.icustay_id
+left join labs_first_day labs
+ on ie.icustay_id = labs.icustay_id
+)
+, scorecomp as
+(
+select
+ cohort.*
+ -- Below code calculates the component scores needed for SAPS
+ , case
+ when age is null then null
+ when age < 40 then 0
+ when age < 60 then 7
+ when age < 70 then 12
+ when age < 75 then 15
+ when age < 80 then 16
+ when age >= 80 then 18
+ end as age_score
+
+ , case
+ when heartrate_max is null then null
+ when heartrate_min < 40 then 11
+ when heartrate_max >= 160 then 7
+ when heartrate_max >= 120 then 4
+ when heartrate_min < 70 then 2
+ when heartrate_max >= 70 and heartrate_max < 120
+ and heartrate_min >= 70 and heartrate_min < 120
+ then 0
+ end as hr_score
+
+ , case
+ when sysbp_min is null then null
+ when sysbp_min < 70 then 13
+ when sysbp_min < 100 then 5
+ when sysbp_max >= 200 then 2
+ when sysbp_max >= 100 and sysbp_max < 200
+ and sysbp_min >= 100 and sysbp_min < 200
+ then 0
+ end as sysbp_score
+
+ , case
+ when tempc_max is null then null
+ when tempc_min < 39.0 then 0
+ when tempc_max >= 39.0 then 3
+ end as temp_score
+
+ , case
+ when pao2fio2_vent_min is null then null
+ when pao2fio2_vent_min < 100 then 11
+ when pao2fio2_vent_min < 200 then 9
+ when pao2fio2_vent_min >= 200 then 6
+ end as pao2fio2_score
+
+ , case
+ when urineoutput is null then null
+ when urineoutput < 500.0 then 11
+ when urineoutput < 1000.0 then 4
+ when urineoutput >= 1000.0 then 0
+ end as uo_score
+
+ , case
+ when bun_max is null then null
+ when bun_max < 28.0 then 0
+ when bun_max < 84.0 then 6
+ when bun_max >= 84.0 then 10
+ end as bun_score
+
+ , case
+ when wbc_max is null then null
+ when wbc_min < 1.0 then 12
+ when wbc_max >= 20.0 then 3
+ when wbc_max >= 1.0 and wbc_max < 20.0
+ and wbc_min >= 1.0 and wbc_min < 20.0
+ then 0
+ end as wbc_score
+
+ , case
+ when potassium_max is null then null
+ when potassium_min < 3.0 then 3
+ when potassium_max >= 5.0 then 3
+ when potassium_max >= 3.0 and potassium_max < 5.0
+ and potassium_min >= 3.0 and potassium_min < 5.0
+ then 0
+ end as potassium_score
+
+ , case
+ when sodium_max is null then null
+ when sodium_min < 125 then 5
+ when sodium_max >= 145 then 1
+ when sodium_max >= 125 and sodium_max < 145
+ and sodium_min >= 125 and sodium_min < 145
+ then 0
+ end as sodium_score
+
+ , case
+ when bicarbonate_max is null then null
+ when bicarbonate_min < 15.0 then 5
+ when bicarbonate_min < 20.0 then 3
+ when bicarbonate_max >= 20.0
+ and bicarbonate_min >= 20.0
+ then 0
+ end as bicarbonate_score
+
+ , case
+ when bilirubin_max is null then null
+ when bilirubin_max < 4.0 then 0
+ when bilirubin_max < 6.0 then 4
+ when bilirubin_max >= 6.0 then 9
+ end as bilirubin_score
+
+ , case
+ when mingcs is null then null
+ when mingcs < 3 then null -- erroneous value/on trach
+ when mingcs < 6 then 26
+ when mingcs < 9 then 13
+ when mingcs < 11 then 7
+ when mingcs < 14 then 5
+ when mingcs >= 14
+ and mingcs <= 15
+ then 0
+ end as gcs_score
+
+ , case
+ when aids = 1 then 17
+ when hem = 1 then 10
+ when mets = 1 then 9
+ else 0
+ end as comorbidity_score
+
+ , case
+ when admissiontype = 'ScheduledSurgical' then 0
+ when admissiontype = 'Medical' then 6
+ when admissiontype = 'UnscheduledSurgical' then 8
+ else null
+ end as admissiontype_score
+
+from cohort
+)
+-- Calculate SAPS II here so we can use it in the probability calculation below
+, score as
+(
+ select s.*
+ -- coalesce statements impute normal score of zero if data element is missing
+ , coalesce(age_score,0)
+ + coalesce(hr_score,0)
+ + coalesce(sysbp_score,0)
+ + coalesce(temp_score,0)
+ + coalesce(pao2fio2_score,0)
+ + coalesce(uo_score,0)
+ + coalesce(bun_score,0)
+ + coalesce(wbc_score,0)
+ + coalesce(potassium_score,0)
+ + coalesce(sodium_score,0)
+ + coalesce(bicarbonate_score,0)
+ + coalesce(bilirubin_score,0)
+ + coalesce(gcs_score,0)
+ + coalesce(comorbidity_score,0)
+ + coalesce(admissiontype_score,0)
+ as sapsii
+ from scorecomp s
+)
+select ie.subject_id, ie.hadm_id, ie.icustay_id
+, sapsii
+, 1 / (1 + exp(- (-7.7631 + 0.0737*(sapsii) + 0.9971*(ln(sapsii + 1))) )) as sapsii_prob
+, age_score
+, hr_score
+, sysbp_score
+, temp_score
+, pao2fio2_score
+, uo_score
+, bun_score
+, wbc_score
+, potassium_score
+, sodium_score
+, bicarbonate_score
+, bilirubin_score
+, gcs_score
+, comorbidity_score
+, admissiontype_score
+from icustays ie
+left join score s
+ on ie.icustay_id = s.icustay_id
+order by ie.icustay_id;
diff --git a/mimic-iii-carevue/code/concepts/46_apsiii.sql b/mimic-iii-carevue/code/concepts/46_apsiii.sql
new file mode 100644
index 000000000..fee02064f
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/46_apsiii.sql
@@ -0,0 +1,835 @@
+drop table if exists apsiii; create table apsiii as
+-- ------------------------------------------------------------------
+-- Title: Acute Physiology Score III (APS III)
+-- This query extracts the acute physiology score III.
+-- This score is a measure of patient severity of illness.
+-- The score is calculated on the first day of each ICU patients' stay.
+-- ------------------------------------------------------------------
+
+-- Reference for APS III:
+-- Knaus WA, Wagner DP, Draper EA, Zimmerman JE, Bergner M, Bastos PG, Sirio CA, Murphy DJ, Lotring T, Damiano A.
+-- The APACHE III prognostic system. Risk prediction of hospital mortality for critically ill hospitalized adults.
+-- Chest Journal. 1991 Dec 1;100(6):1619-36.
+
+-- Reference for the equation for calibrating APS III to hospital mortality:
+-- Johnson, A. E. W. (2015). Mortality prediction and acuity assessment in critical care.
+-- University of Oxford, Oxford, UK.
+
+-- Variables used in APS III:
+-- GCS
+-- VITALS: Heart rate, mean blood pressure, temperature, respiration rate
+-- FLAGS: ventilation/cpap, chronic dialysis
+-- IO: urine output
+-- LABS: pao2, A-aDO2, hematocrit, WBC, creatinine
+-- , blood urea nitrogen, sodium, albumin, bilirubin, glucose, pH, pCO2
+
+-- The following views are required to run this query:
+-- 1) urine_output_first_day - generated by urine-output-first-day.sql
+-- 2) ventilation_first_day - generated by ventilated-first-day.sql
+-- 3) vitals_first_day - generated by vitals-first-day.sql
+-- 4) gcs_first_day - generated by gcs-first-day.sql
+-- 5) labs_first_day - generated by labs-first-day.sql
+
+-- Note:
+-- The score is calculated for *all* ICU patients, with the assumption that the user will subselect appropriate icustay_ids.
+-- For example, the score is calculated for neonates, but it is likely inappropriate to actually use the score values for these patients.
+
+-- List of TODO:
+-- The site of temperature is not incorporated. Axillary measurements should be increased by 1 degree.
+-- Unfortunately the data for metavision is not available at the moment.
+-- 674 | Temp. Site
+-- 224642 | Temperature Site
+
+with pa as
+(
+ select bg.icustay_id, bg.charttime
+ , po2 as PaO2
+ , row_number() over (partition by bg.icustay_id order by bg.po2 desc) as rn
+ from blood_gas_first_day_arterial bg
+ left join ventilation_durations vd
+ on bg.icustay_id = vd.icustay_id
+ and bg.charttime >= vd.starttime
+ and bg.charttime <= vd.endtime
+ where vd.icustay_id is null -- patient is *not* ventilated
+ -- and fio2 < 50, or if no fio2, assume room air
+ and coalesce(fio2, fio2_chartevents, 21) < 50
+ and bg.po2 is not null
+)
+, aa as
+(
+ -- join blood gas to ventilation durations to determine if patient was vent
+ -- also join to cpap table for the same purpose
+ select bg.icustay_id, bg.charttime
+ , bg.aado2
+ , row_number() over (partition by bg.icustay_id order by bg.aado2 desc) as rn
+ -- row number indicating the highest AaDO2
+ from blood_gas_first_day_arterial bg
+ inner join ventilation_durations vd
+ on bg.icustay_id = vd.icustay_id
+ and bg.charttime >= vd.starttime
+ and bg.charttime <= vd.endtime
+ where vd.icustay_id is not null -- patient is ventilated
+ and coalesce(fio2, fio2_chartevents) >= 50
+ and bg.aado2 is not null
+)
+-- because ph/pco2 rules are an interaction *within* a blood gas, we calculate them here
+-- the worse score is then taken for the final calculation
+, acidbase as
+(
+ select bg.icustay_id
+ , ph, pco2 as paco2
+ , case
+ when ph is null or pco2 is null then null
+ when ph < 7.20 then
+ case
+ when pco2 < 50 then 12
+ else 4
+ end
+ when ph < 7.30 then
+ case
+ when pco2 < 30 then 9
+ when pco2 < 40 then 6
+ when pco2 < 50 then 3
+ else 2
+ end
+ when ph < 7.35 then
+ case
+ when pco2 < 30 then 9
+ when pco2 < 45 then 0
+ else 1
+ end
+ when ph < 7.45 then
+ case
+ when pco2 < 30 then 5
+ when pco2 < 45 then 0
+ else 1
+ end
+ when ph < 7.50 then
+ case
+ when pco2 < 30 then 5
+ when pco2 < 35 then 0
+ when pco2 < 45 then 2
+ else 12
+ end
+ when ph < 7.60 then
+ case
+ when pco2 < 40 then 3
+ else 12
+ end
+ else -- ph >= 7.60
+ case
+ when pco2 < 25 then 0
+ when pco2 < 40 then 3
+ else 12
+ end
+ end as acidbase_score
+ from blood_gas_first_day_arterial bg
+ where ph is not null and pco2 is not null
+)
+, acidbase_max as
+(
+ select icustay_id, acidbase_score, ph, paco2
+ -- create integer which indexes maximum value of score with 1
+ , row_number() over (partition by icustay_id order by acidbase_score desc) as acidbase_rn
+ from acidbase
+)
+-- define acute renal failure (ARF) as:
+-- creatinine >=1.5 mg/dl
+-- and urine output <410 cc/day
+-- and no chronic dialysis
+, arf as
+(
+ select ie.icustay_id
+ , case
+ when labs.creatinine_max >= 1.5
+ and uo.urineoutput < 410
+ -- acute renal failure is only coded if the patient is not on chronic dialysis
+ -- we use ICD-9 coding of ESRD as a proxy for chronic dialysis
+ and icd.ckd = 0
+ then 1
+ else 0 end as arf
+ from icustays ie
+ left join urine_output_first_day uo
+ on ie.icustay_id = uo.icustay_id
+ left join labs_first_day labs
+ on ie.icustay_id = labs.icustay_id
+ left join
+ (
+ select hadm_id
+ , max(case
+ -- severe kidney failure requiring use of dialysis
+ when icd9_code in ('5854','5855','5856') then 1
+ -- we do not include 5859 as that is sometimes coded for acute-on-chronic ARF
+ else 0 end)
+ as ckd
+ from diagnoses_icd
+ group by hadm_id
+ ) icd
+ on ie.hadm_id = icd.hadm_id
+)
+, cohort as
+(
+select ie.subject_id, ie.hadm_id, ie.icustay_id
+ , ie.intime
+ , ie.outtime
+
+ , vital.heartrate_min
+ , vital.heartrate_max
+ , vital.meanbp_min
+ , vital.meanbp_max
+ , vital.tempc_min
+ , vital.tempc_max
+ , vital.resprate_min
+ , vital.resprate_max
+
+ , pa.pao2
+ , aa.aado2
+
+ , ab.ph
+ , ab.paco2
+ , ab.acidbase_score
+
+ , labs.hematocrit_min
+ , labs.hematocrit_max
+ , labs.wbc_min
+ , labs.wbc_max
+ , labs.creatinine_min
+ , labs.creatinine_max
+ , labs.bun_min
+ , labs.bun_max
+ , labs.sodium_min
+ , labs.sodium_max
+ , labs.albumin_min
+ , labs.albumin_max
+ , labs.bilirubin_min
+ , labs.bilirubin_max
+
+ , case
+ when labs.glucose_max is null and vital.glucose_max is null
+ then null
+ when labs.glucose_max is null or vital.glucose_max > labs.glucose_max
+ then vital.glucose_max
+ when vital.glucose_max is null or labs.glucose_max > vital.glucose_max
+ then labs.glucose_max
+ else labs.glucose_max -- if equal, just pick labs
+ end as glucose_max
+
+ , case
+ when labs.glucose_min is null and vital.glucose_min is null
+ then null
+ when labs.glucose_min is null or vital.glucose_min < labs.glucose_min
+ then vital.glucose_min
+ when vital.glucose_min is null or labs.glucose_min < vital.glucose_min
+ then labs.glucose_min
+ else labs.glucose_min -- if equal, just pick labs
+ end as glucose_min
+
+ -- , labs.bicarbonate_min
+ -- , labs.bicarbonate_max
+ , vent.vent
+ , uo.urineoutput
+ -- gcs and its components
+ , gcs.mingcs
+ , gcs.gcsmotor, gcs.gcsverbal, gcs.gcseyes, gcs.endotrachflag
+ -- acute renal failure
+ , arf.arf as arf
+
+from icustays ie
+inner join admissions adm
+ on ie.hadm_id = adm.hadm_id
+inner join patients pat
+ on ie.subject_id = pat.subject_id
+
+-- join to above views - the row number filters to 1 row per icustay_id
+left join pa
+ on ie.icustay_id = pa.icustay_id
+ and pa.rn = 1
+left join aa
+ on ie.icustay_id = aa.icustay_id
+ and aa.rn = 1
+left join acidbase_max ab
+ on ie.icustay_id = ab.icustay_id
+ and ab.acidbase_rn = 1
+left join arf
+ on ie.icustay_id = arf.icustay_id
+
+-- join to custom tables to get more data....
+left join ventilation_first_day vent
+ on ie.icustay_id = vent.icustay_id
+left join gcs_first_day gcs
+ on ie.icustay_id = gcs.icustay_id
+left join vitals_first_day vital
+ on ie.icustay_id = vital.icustay_id
+left join urine_output_first_day uo
+ on ie.icustay_id = uo.icustay_id
+left join labs_first_day labs
+ on ie.icustay_id = labs.icustay_id
+)
+-- First, we calculate the score for the minimum values
+, score_min as
+(
+ select cohort.subject_id, cohort.hadm_id, cohort.icustay_id
+ , case
+ when heartrate_min is null then null
+ when heartrate_min < 40 then 8
+ when heartrate_min < 50 then 5
+ when heartrate_min < 100 then 0
+ when heartrate_min < 110 then 1
+ when heartrate_min < 120 then 5
+ when heartrate_min < 140 then 7
+ when heartrate_min < 155 then 13
+ when heartrate_min >= 155 then 17
+ end as hr_score
+
+ , case
+ when meanbp_min is null then null
+ when meanbp_min < 40 then 23
+ when meanbp_min < 60 then 15
+ when meanbp_min < 70 then 7
+ when meanbp_min < 80 then 6
+ when meanbp_min < 100 then 0
+ when meanbp_min < 120 then 4
+ when meanbp_min < 130 then 7
+ when meanbp_min < 140 then 9
+ when meanbp_min >= 140 then 10
+ end as meanbp_score
+
+ -- TODO: add 1 degree to axillary measurements
+ , case
+ when tempc_min is null then null
+ when tempc_min < 33.0 then 20
+ when tempc_min < 33.5 then 16
+ when tempc_min < 34.0 then 13
+ when tempc_min < 35.0 then 8
+ when tempc_min < 36.0 then 2
+ when tempc_min < 40.0 then 0
+ when tempc_min >= 40.0 then 4
+ end as temp_score
+
+ , case
+ when resprate_min is null then null
+ -- special case for ventilated patients
+ when vent = 1 and resprate_min < 14 then 0
+ when resprate_min < 6 then 17
+ when resprate_min < 12 then 8
+ when resprate_min < 14 then 7
+ when resprate_min < 25 then 0
+ when resprate_min < 35 then 6
+ when resprate_min < 40 then 9
+ when resprate_min < 50 then 11
+ when resprate_min >= 50 then 18
+ end as resprate_score
+
+ , case
+ when hematocrit_min is null then null
+ when hematocrit_min < 41.0 then 3
+ when hematocrit_min < 50.0 then 0
+ when hematocrit_min >= 50.0 then 3
+ end as hematocrit_score
+
+ , case
+ when wbc_min is null then null
+ when wbc_min < 1.0 then 19
+ when wbc_min < 3.0 then 5
+ when wbc_min < 20.0 then 0
+ when wbc_min < 25.0 then 1
+ when wbc_min >= 25.0 then 5
+ end as wbc_score
+
+ , case
+ when creatinine_min is null then null
+ when arf = 1 and creatinine_min < 1.5 then 0
+ when arf = 1 and creatinine_min >= 1.5 then 10
+ when creatinine_min < 0.5 then 3
+ when creatinine_min < 1.5 then 0
+ when creatinine_min < 1.95 then 4
+ when creatinine_min >= 1.95 then 7
+ end as creatinine_score
+
+ , case
+ when bun_min is null then null
+ when bun_min < 17.0 then 0
+ when bun_min < 20.0 then 2
+ when bun_min < 40.0 then 7
+ when bun_min < 80.0 then 11
+ when bun_min >= 80.0 then 12
+ end as bun_score
+
+ , case
+ when sodium_min is null then null
+ when sodium_min < 120 then 3
+ when sodium_min < 135 then 2
+ when sodium_min < 155 then 0
+ when sodium_min >= 155 then 4
+ end as sodium_score
+
+ , case
+ when albumin_min is null then null
+ when albumin_min < 2.0 then 11
+ when albumin_min < 2.5 then 6
+ when albumin_min < 4.5 then 0
+ when albumin_min >= 4.5 then 4
+ end as albumin_score
+
+ , case
+ when bilirubin_min is null then null
+ when bilirubin_min < 2.0 then 0
+ when bilirubin_min < 3.0 then 5
+ when bilirubin_min < 5.0 then 6
+ when bilirubin_min < 8.0 then 8
+ when bilirubin_min >= 8.0 then 16
+ end as bilirubin_score
+
+ , case
+ when glucose_min is null then null
+ when glucose_min < 40 then 8
+ when glucose_min < 60 then 9
+ when glucose_min < 200 then 0
+ when glucose_min < 350 then 3
+ when glucose_min >= 350 then 5
+ end as glucose_score
+
+from cohort
+)
+, score_max as
+(
+ select cohort.subject_id, cohort.hadm_id, cohort.icustay_id
+ , case
+ when heartrate_max is null then null
+ when heartrate_max < 40 then 8
+ when heartrate_max < 50 then 5
+ when heartrate_max < 100 then 0
+ when heartrate_max < 110 then 1
+ when heartrate_max < 120 then 5
+ when heartrate_max < 140 then 7
+ when heartrate_max < 155 then 13
+ when heartrate_max >= 155 then 17
+ end as hr_score
+
+ , case
+ when meanbp_max is null then null
+ when meanbp_max < 40 then 23
+ when meanbp_max < 60 then 15
+ when meanbp_max < 70 then 7
+ when meanbp_max < 80 then 6
+ when meanbp_max < 100 then 0
+ when meanbp_max < 120 then 4
+ when meanbp_max < 130 then 7
+ when meanbp_max < 140 then 9
+ when meanbp_max >= 140 then 10
+ end as meanbp_score
+
+ -- TODO: add 1 degree to axillary measurements
+ , case
+ when tempc_max is null then null
+ when tempc_max < 33.0 then 20
+ when tempc_max < 33.5 then 16
+ when tempc_max < 34.0 then 13
+ when tempc_max < 35.0 then 8
+ when tempc_max < 36.0 then 2
+ when tempc_max < 40.0 then 0
+ when tempc_max >= 40.0 then 4
+ end as temp_score
+
+ , case
+ when resprate_max is null then null
+ -- special case for ventilated patients
+ when vent = 1 and resprate_max < 14 then 0
+ when resprate_max < 6 then 17
+ when resprate_max < 12 then 8
+ when resprate_max < 14 then 7
+ when resprate_max < 25 then 0
+ when resprate_max < 35 then 6
+ when resprate_max < 40 then 9
+ when resprate_max < 50 then 11
+ when resprate_max >= 50 then 18
+ end as resprate_score
+
+ , case
+ when hematocrit_max is null then null
+ when hematocrit_max < 41.0 then 3
+ when hematocrit_max < 50.0 then 0
+ when hematocrit_max >= 50.0 then 3
+ end as hematocrit_score
+
+ , case
+ when wbc_max is null then null
+ when wbc_max < 1.0 then 19
+ when wbc_max < 3.0 then 5
+ when wbc_max < 20.0 then 0
+ when wbc_max < 25.0 then 1
+ when wbc_max >= 25.0 then 5
+ end as wbc_score
+
+ , case
+ when creatinine_max is null then null
+ when arf = 1 and creatinine_max < 1.5 then 0
+ when arf = 1 and creatinine_max >= 1.5 then 10
+ when creatinine_max < 0.5 then 3
+ when creatinine_max < 1.5 then 0
+ when creatinine_max < 1.95 then 4
+ when creatinine_max >= 1.95 then 7
+ end as creatinine_score
+
+ , case
+ when bun_max is null then null
+ when bun_max < 17.0 then 0
+ when bun_max < 20.0 then 2
+ when bun_max < 40.0 then 7
+ when bun_max < 80.0 then 11
+ when bun_max >= 80.0 then 12
+ end as bun_score
+
+ , case
+ when sodium_max is null then null
+ when sodium_max < 120 then 3
+ when sodium_max < 135 then 2
+ when sodium_max < 155 then 0
+ when sodium_max >= 155 then 4
+ end as sodium_score
+
+ , case
+ when albumin_max is null then null
+ when albumin_max < 2.0 then 11
+ when albumin_max < 2.5 then 6
+ when albumin_max < 4.5 then 0
+ when albumin_max >= 4.5 then 4
+ end as albumin_score
+
+ , case
+ when bilirubin_max is null then null
+ when bilirubin_max < 2.0 then 0
+ when bilirubin_max < 3.0 then 5
+ when bilirubin_max < 5.0 then 6
+ when bilirubin_max < 8.0 then 8
+ when bilirubin_max >= 8.0 then 16
+ end as bilirubin_score
+
+ , case
+ when glucose_max is null then null
+ when glucose_max < 40 then 8
+ when glucose_max < 60 then 9
+ when glucose_max < 200 then 0
+ when glucose_max < 350 then 3
+ when glucose_max >= 350 then 5
+ end as glucose_score
+
+from cohort
+)
+-- Combine together the scores for min/max, using the following rules:
+-- 1) select the value furthest from a predefined normal value
+-- 2) if both equidistant, choose the one which gives a worse score
+-- 3) calculate score for acid-base abnormalities as it requires interactions
+-- sometimes the code is a bit redundant, i.e. we know the max would always be furthest from 0
+, scorecomp as
+(
+ select co.*
+ -- The rules for APS III require the definition of a "worst" value
+ -- This value is defined as whatever value is furthest from a predefined normal
+ -- e.g., for heart rate, worst is defined as furthest from 75
+ , case
+ when heartrate_max is null then null
+ when abs(heartrate_max-75) > abs(heartrate_min-75)
+ then smax.hr_score
+ when abs(heartrate_max-75) < abs(heartrate_min-75)
+ then smin.hr_score
+ when abs(heartrate_max-75) = abs(heartrate_min-75)
+ and smax.hr_score >= smin.hr_score
+ then smax.hr_score
+ when abs(heartrate_max-75) = abs(heartrate_min-75)
+ and smax.hr_score < smin.hr_score
+ then smin.hr_score
+ end as hr_score
+
+ , case
+ when meanbp_max is null then null
+ when abs(meanbp_max-90) > abs(meanbp_min-90)
+ then smax.meanbp_score
+ when abs(meanbp_max-90) < abs(meanbp_min-90)
+ then smin.meanbp_score
+ -- values are equidistant - pick the larger score
+ when abs(meanbp_max-90) = abs(meanbp_min-90)
+ and smax.meanbp_score >= smin.meanbp_score
+ then smax.meanbp_score
+ when abs(meanbp_max-90) = abs(meanbp_min-90)
+ and smax.meanbp_score < smin.meanbp_score
+ then smin.meanbp_score
+ end as meanbp_score
+
+ , case
+ when tempc_max is null then null
+ when abs(tempc_max-38) > abs(tempc_min-38)
+ then smax.temp_score
+ when abs(tempc_max-38) < abs(tempc_min-38)
+ then smin.temp_score
+ -- values are equidistant - pick the larger score
+ when abs(tempc_max-38) = abs(tempc_min-38)
+ and smax.temp_score >= smin.temp_score
+ then smax.temp_score
+ when abs(tempc_max-38) = abs(tempc_min-38)
+ and smax.temp_score < smin.temp_score
+ then smin.temp_score
+ end as temp_score
+
+ , case
+ when resprate_max is null then null
+ when abs(resprate_max-19) > abs(resprate_min-19)
+ then smax.resprate_score
+ when abs(resprate_max-19) < abs(resprate_min-19)
+ then smin.resprate_score
+ -- values are equidistant - pick the larger score
+ when abs(resprate_max-19) = abs(resprate_max-19)
+ and smax.resprate_score >= smin.resprate_score
+ then smax.resprate_score
+ when abs(resprate_max-19) = abs(resprate_max-19)
+ and smax.resprate_score < smin.resprate_score
+ then smin.resprate_score
+ end as resprate_score
+
+ , case
+ when hematocrit_max is null then null
+ when abs(hematocrit_max-45.5) > abs(hematocrit_min-45.5)
+ then smax.hematocrit_score
+ when abs(hematocrit_max-45.5) < abs(hematocrit_min-45.5)
+ then smin.hematocrit_score
+ -- values are equidistant - pick the larger score
+ when abs(hematocrit_max-45.5) = abs(hematocrit_max-45.5)
+ and smax.hematocrit_score >= smin.hematocrit_score
+ then smax.hematocrit_score
+ when abs(hematocrit_max-45.5) = abs(hematocrit_max-45.5)
+ and smax.hematocrit_score < smin.hematocrit_score
+ then smin.hematocrit_score
+ end as hematocrit_score
+
+ , case
+ when wbc_max is null then null
+ when abs(wbc_max-11.5) > abs(wbc_min-11.5)
+ then smax.wbc_score
+ when abs(wbc_max-11.5) < abs(wbc_min-11.5)
+ then smin.wbc_score
+ -- values are equidistant - pick the larger score
+ when abs(wbc_max-11.5) = abs(wbc_max-11.5)
+ and smax.wbc_score >= smin.wbc_score
+ then smax.wbc_score
+ when abs(wbc_max-11.5) = abs(wbc_max-11.5)
+ and smax.wbc_score < smin.wbc_score
+ then smin.wbc_score
+ end as wbc_score
+
+
+ -- For some labs, "furthest from normal" doesn't make sense
+ -- e.g. creatinine w/ ARF, the minimum could be 0.3, and the max 1.6
+ -- while the minimum of 0.3 is "further from 1", seems like the max should be scored
+
+ , case
+ when creatinine_max is null then null
+ -- if they have arf then use the max to score
+ when arf = 1 then smax.creatinine_score
+ -- otherwise furthest from 1
+ when abs(creatinine_max-1) > abs(creatinine_min-1)
+ then smax.creatinine_score
+ when abs(creatinine_max-1) < abs(creatinine_min-1)
+ then smin.creatinine_score
+ -- values are equidistant
+ when smax.creatinine_score >= smin.creatinine_score
+ then smax.creatinine_score
+ when smax.creatinine_score < smin.creatinine_score
+ then smin.creatinine_score
+ end as creatinine_score
+
+ -- the rule for BUN is the furthest from 0.. equivalent to the max value
+ , case
+ when bun_max is null then null
+ else smax.bun_score
+ end as bun_score
+
+ , case
+ when sodium_max is null then null
+ when abs(sodium_max-145.5) > abs(sodium_min-145.5)
+ then smax.sodium_score
+ when abs(sodium_max-145.5) < abs(sodium_min-145.5)
+ then smin.sodium_score
+ -- values are equidistant - pick the larger score
+ when abs(sodium_max-145.5) = abs(sodium_max-145.5)
+ and smax.sodium_score >= smin.sodium_score
+ then smax.sodium_score
+ when abs(sodium_max-145.5) = abs(sodium_max-145.5)
+ and smax.sodium_score < smin.sodium_score
+ then smin.sodium_score
+ end as sodium_score
+
+ , case
+ when albumin_max is null then null
+ when abs(albumin_max-3.5) > abs(albumin_min-3.5)
+ then smax.albumin_score
+ when abs(albumin_max-3.5) < abs(albumin_min-3.5)
+ then smin.albumin_score
+ -- values are equidistant - pick the larger score
+ when abs(albumin_max-3.5) = abs(albumin_max-3.5)
+ and smax.albumin_score >= smin.albumin_score
+ then smax.albumin_score
+ when abs(albumin_max-3.5) = abs(albumin_max-3.5)
+ and smax.albumin_score < smin.albumin_score
+ then smin.albumin_score
+ end as albumin_score
+
+ , case
+ when bilirubin_max is null then null
+ else smax.bilirubin_score
+ end as bilirubin_score
+
+ , case
+ when glucose_max is null then null
+ when abs(glucose_max-130) > abs(glucose_min-130)
+ then smax.glucose_score
+ when abs(glucose_max-130) < abs(glucose_min-130)
+ then smin.glucose_score
+ -- values are equidistant - pick the larger score
+ when abs(glucose_max-130) = abs(glucose_max-130)
+ and smax.glucose_score >= smin.glucose_score
+ then smax.glucose_score
+ when abs(glucose_max-130) = abs(glucose_max-130)
+ and smax.glucose_score < smin.glucose_score
+ then smin.glucose_score
+ end as glucose_score
+
+
+ -- Below are interactions/special cases where only 1 value is important
+ , case
+ when urineoutput is null then null
+ when urineoutput < 400 then 15
+ when urineoutput < 600 then 8
+ when urineoutput < 900 then 7
+ when urineoutput < 1500 then 5
+ when urineoutput < 2000 then 4
+ when urineoutput < 4000 then 0
+ when urineoutput >= 4000 then 1
+ end as uo_score
+
+ , case
+ when endotrachflag = 1
+ -- here they are intubated, so their verbal score is inappropriate
+ -- normally you are supposed to use "clinical judgement"
+ -- we don't have that, so we just assume normal (as was done in the original study)
+ then 0
+ when gcseyes = 1
+ then case
+ when gcsverbal = 1 and gcsmotor in (1,2)
+ then 48
+ when gcsverbal = 1 and gcsmotor in (3,4)
+ then 33
+ when gcsverbal = 1 and gcsmotor in (5,6)
+ then 16
+ when gcsverbal in (2,3) and gcsmotor in (1,2)
+ then 29
+ when gcsverbal in (2,3) and gcsmotor in (3,4)
+ then 24
+ when gcsverbal in (2,3) and gcsmotor >= 5
+ -- highly unlikely clinical combination
+ then null
+ when gcsverbal >= 4
+ then null
+ end
+ when gcseyes > 1
+ then case
+ when gcsverbal = 1 and gcsmotor in (1,2)
+ then 29
+ when gcsverbal = 1 and gcsmotor in (3,4)
+ then 24
+ when gcsverbal = 1 and gcsmotor in (5,6)
+ then 15
+ when gcsverbal in (2,3) and gcsmotor in (1,2)
+ then 29
+ when gcsverbal in (2,3) and gcsmotor in (3,4)
+ then 24
+ when gcsverbal in (2,3) and gcsmotor = 5
+ then 13
+ when gcsverbal in (2,3) and gcsmotor = 6
+ then 10
+ when gcsverbal = 4 and gcsmotor in (1,2,3,4)
+ then 13
+ when gcsverbal = 4 and gcsmotor = 5
+ then 8
+ when gcsverbal = 4 and gcsmotor = 6
+ then 3
+ when gcsverbal = 5 and gcsmotor in (1,2,3,4,5)
+ then 3
+ when gcsverbal = 5 and gcsmotor = 6
+ then 0
+ end
+ else null
+ end as gcs_score
+
+ , case
+ when pao2 is null and aado2 is null
+ then null
+ when pao2 is not null then
+ case
+ when pao2 < 50 then 15
+ when pao2 < 70 then 5
+ when pao2 < 80 then 2
+ else 0 end
+ when aado2 is not null then
+ case
+ when aado2 < 100 then 0
+ when aado2 < 250 then 7
+ when aado2 < 350 then 9
+ when aado2 < 500 then 11
+ when aado2 >= 500 then 14
+ else 0 end
+ end as pao2_aado2_score
+
+from cohort co
+left join score_min smin
+ on co.icustay_id = smin.icustay_id
+left join score_max smax
+ on co.icustay_id = smax.icustay_id
+)
+-- tabulate the APS III using the scores from the worst values
+, score as
+(
+ select s.*
+ -- coalesce statements impute normal score of zero if data element is missing
+ , coalesce(hr_score,0)
+ + coalesce(meanbp_score,0)
+ + coalesce(temp_score,0)
+ + coalesce(resprate_score,0)
+ + coalesce(pao2_aado2_score,0)
+ + coalesce(hematocrit_score,0)
+ + coalesce(wbc_score,0)
+ + coalesce(creatinine_score,0)
+ + coalesce(uo_score,0)
+ + coalesce(bun_score,0)
+ + coalesce(sodium_score,0)
+ + coalesce(albumin_score,0)
+ + coalesce(bilirubin_score,0)
+ + coalesce(glucose_score,0)
+ + coalesce(acidbase_score,0)
+ + coalesce(gcs_score,0)
+ as apsiii
+ from scorecomp s
+)
+select ie.subject_id, ie.hadm_id, ie.icustay_id
+, apsiii
+-- Calculate probability of hospital mortality using equation from Johnson 2014.
+, 1 / (1 + exp(- (-4.4360 + 0.04726*(apsiii) ))) as apsiii_prob
+, hr_score
+, meanbp_score
+, temp_score
+, resprate_score
+, pao2_aado2_score
+, hematocrit_score
+, wbc_score
+, creatinine_score
+, uo_score
+, bun_score
+, sodium_score
+, albumin_score
+, bilirubin_score
+, glucose_score
+, acidbase_score
+, gcs_score
+from icustays ie
+left join score s
+ on ie.icustay_id = s.icustay_id
+order by ie.icustay_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/47_lods.sql b/mimic-iii-carevue/code/concepts/47_lods.sql
new file mode 100644
index 000000000..774841f3d
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/47_lods.sql
@@ -0,0 +1,235 @@
+drop table if exists lods; create table lods as
+-- ------------------------------------------------------------------
+-- Title: Logistic Organ Dysfunction Score (LODS)
+-- This query extracts the logistic organ dysfunction system.
+-- This score is a measure of organ failure in a patient.
+-- The score is calculated on the first day of each ICU patients' stay.
+-- ------------------------------------------------------------------
+
+-- Reference for LODS:
+-- Le Gall, J. R., Klar, J., Lemeshow, S., Saulnier, F., Alberti, C., Artigas, A., & Teres, D.
+-- The Logistic Organ Dysfunction system: a new way to assess organ dysfunction in the intensive care unit.
+-- JAMA 276.10 (1996): 802-810.
+
+-- Variables used in LODS:
+-- GCS
+-- VITALS: Heart rate, systolic blood pressure
+-- FLAGS: ventilation/cpap
+-- IO: urine output
+-- LABS: blood urea nitrogen, WBC, bilirubin, creatinine, prothrombin time (PT), platelets
+-- ABG: PaO2 with associated FiO2
+
+-- The following views are required to run this query:
+-- 1) urine_output_first_day - generated by urine-output-first-day.sql
+-- 2) ventilation_durations - generated by ventilation_durations.sql
+-- 3) vitals_first_day - generated by vitals-first-day.sql
+-- 4) gcs_first_day - generated by gcs-first-day.sql
+-- 5) labs_first_day - generated by labs-first-day.sql
+-- 5) blood_gas_first_day_arterial - generated by blood-gas-first-day-arterial.sql
+
+-- Note:
+-- The score is calculated for *all* ICU patients, with the assumption that the user will subselect appropriate ICUSTAY_IDs.
+-- For example, the score is calculated for neonates, but it is likely inappropriate to actually use the score values for these patients.
+
+-- extract CPAP from the "Oxygen Delivery Device" fields
+with cpap as
+(
+ select ie.icustay_id
+ , min(charttime - interval '1 hour') as starttime
+ , max(charttime + interval '4 hour') as endtime
+ , max(case
+ when lower(ce.value) like '%cpap%' then 1
+ when lower(ce.value) like '%bipap mask%' then 1
+ else 0 end) as cpap
+ from icustays ie
+ inner join chartevents ce
+ on ie.icustay_id = ce.icustay_id
+ and ce.charttime between ie.intime and (ie.intime + interval '1 day')
+ where itemid in
+ (
+ -- TODO: when metavision data import fixed, check the values in 226732 match the value clause below
+ 467, 469, 226732
+ )
+ and (lower(ce.value) like '%cpap%' or lower(ce.value) like '%bipap mask%')
+ -- exclude rows marked as error
+ and (ce.error is null or ce.error = 0)
+ group by ie.icustay_id
+)
+, pafi1 as
+(
+ -- join blood gas to ventilation durations to determine if patient was vent
+ -- also join to cpap table for the same purpose
+ select bg.icustay_id, bg.charttime
+ , pao2fio2
+ , case when vd.icustay_id is not null then 1 else 0 end as vent
+ , case when cp.icustay_id is not null then 1 else 0 end as cpap
+ from blood_gas_first_day_arterial bg
+ left join ventilation_durations vd
+ on bg.icustay_id = vd.icustay_id
+ and bg.charttime >= vd.starttime
+ and bg.charttime <= vd.endtime
+ left join cpap cp
+ on bg.icustay_id = cp.icustay_id
+ and bg.charttime >= cp.starttime
+ and bg.charttime <= cp.endtime
+)
+, pafi2 as
+(
+ -- get the minimum PaO2/FiO2 ratio *only for ventilated/cpap patients*
+ select icustay_id
+ , min(pao2fio2) as pao2fio2_vent_min
+ from pafi1
+ where vent = 1 or cpap = 1
+ group by icustay_id
+)
+, cohort as
+(
+select ie.subject_id
+ , ie.hadm_id
+ , ie.icustay_id
+ , ie.intime
+ , ie.outtime
+
+ , gcs.mingcs
+ , vital.heartrate_max
+ , vital.heartrate_min
+ , vital.sysbp_max
+ , vital.sysbp_min
+
+ -- this value is non-null iff the patient is on vent/cpap
+ , pf.pao2fio2_vent_min
+
+ , labs.bun_max
+ , labs.bun_min
+ , labs.wbc_max
+ , labs.wbc_min
+ , labs.bilirubin_max
+ , labs.creatinine_max
+ , labs.pt_min
+ , labs.pt_max
+ , labs.platelet_min
+
+ , uo.urineoutput
+
+from icustays ie
+inner join admissions adm
+ on ie.hadm_id = adm.hadm_id
+inner join patients pat
+ on ie.subject_id = pat.subject_id
+
+-- join to above view to get pao2/fio2 ratio
+left join pafi2 pf
+ on ie.icustay_id = pf.icustay_id
+
+-- join to custom tables to get more data....
+left join gcs_first_day gcs
+ on ie.icustay_id = gcs.icustay_id
+left join vitals_first_day vital
+ on ie.icustay_id = vital.icustay_id
+left join urine_output_first_day uo
+ on ie.icustay_id = uo.icustay_id
+left join labs_first_day labs
+ on ie.icustay_id = labs.icustay_id
+)
+, scorecomp as
+(
+select
+ cohort.*
+ -- Below code calculates the component scores needed for SAPS
+
+ -- neurologic
+ , case
+ when mingcs is null then null
+ when mingcs < 3 then null -- erroneous value/on trach
+ when mingcs <= 5 then 5
+ when mingcs <= 8 then 3
+ when mingcs <= 13 then 1
+ else 0
+ end as neurologic
+
+ -- cardiovascular
+ , case
+ when heartrate_max is null
+ and sysbp_min is null then null
+ when heartrate_min < 30 then 5
+ when sysbp_min < 40 then 5
+ when sysbp_min < 70 then 3
+ when sysbp_max >= 270 then 3
+ when heartrate_max >= 140 then 1
+ when sysbp_max >= 240 then 1
+ when sysbp_min < 90 then 1
+ else 0
+ end as cardiovascular
+
+ -- renal
+ , case
+ when bun_max is null
+ or urineoutput is null
+ or creatinine_max is null
+ then null
+ when urineoutput < 500.0 then 5
+ when bun_max >= 56.0 then 5
+ when creatinine_max >= 1.60 then 3
+ when urineoutput < 750.0 then 3
+ when bun_max >= 28.0 then 3
+ when urineoutput >= 10000.0 then 3
+ when creatinine_max >= 1.20 then 1
+ when bun_max >= 17.0 then 1
+ when bun_max >= 7.50 then 1
+ else 0
+ end as renal
+
+ -- pulmonary
+ , case
+ when pao2fio2_vent_min is null then 0
+ when pao2fio2_vent_min >= 150 then 1
+ when pao2fio2_vent_min < 150 then 3
+ else null
+ end as pulmonary
+
+ -- hematologic
+ , case
+ when wbc_max is null
+ and platelet_min is null
+ then null
+ when wbc_min < 1.0 then 3
+ when wbc_min < 2.5 then 1
+ when platelet_min < 50.0 then 1
+ when wbc_max >= 50.0 then 1
+ else 0
+ end as hematologic
+
+ -- hepatic
+ -- We have defined the "standard" PT as 12 seconds.
+ -- This is an assumption and subsequent analyses may be affected by this assumption.
+ , case
+ when pt_max is null
+ and bilirubin_max is null
+ then null
+ when bilirubin_max >= 2.0 then 1
+ when pt_max > (12+3) then 1
+ when pt_min < (12*0.25) then 1
+ else 0
+ end as hepatic
+
+from cohort
+)
+select ie.subject_id, ie.hadm_id, ie.icustay_id
+-- coalesce statements impute normal score of zero if data element is missing
+, coalesce(neurologic,0)
++ coalesce(cardiovascular,0)
++ coalesce(renal,0)
++ coalesce(pulmonary,0)
++ coalesce(hematologic,0)
++ coalesce(hepatic,0)
+ as lods
+, neurologic
+, cardiovascular
+, renal
+, pulmonary
+, hematologic
+, hepatic
+from icustays ie
+left join scorecomp s
+ on ie.icustay_id = s.icustay_id
+order by ie.icustay_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/48_sirs.sql b/mimic-iii-carevue/code/concepts/48_sirs.sql
new file mode 100644
index 000000000..c472635f0
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/48_sirs.sql
@@ -0,0 +1,112 @@
+drop table if exists sirs; create table sirs as
+-- ------------------------------------------------------------------
+-- Title: Systemic inflammatory response syndrome (SIRS) criteria
+-- This query extracts the Systemic inflammatory response syndrome (SIRS) criteria
+-- The criteria quantify the level of inflammatory response of the body
+-- The score is calculated on the first day of each ICU patients' stay.
+-- ------------------------------------------------------------------
+
+-- Reference for SIRS:
+-- American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference:
+-- definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis"
+-- Crit. Care Med. 20 (6): 864–74. 1992.
+-- doi:10.1097/00003246-199206000-00025. PMID 1597042.
+
+-- Variables used in SIRS:
+-- Body temperature (min and max)
+-- Heart rate (max)
+-- Respiratory rate (max)
+-- PaCO2 (min)
+-- White blood cell count (min and max)
+-- the presence of greater than 10% immature neutrophils (band forms)
+
+-- The following views required to run this query:
+-- 1) vitals_first_day - generated by vitals-first-day.sql
+-- 2) labs_first_day - generated by labs-first-day.sql
+-- 3) blood_gas_first_day_arterial - generated by blood-gas-first-day-arterial.sql
+
+-- Note:
+-- The score is calculated for *all* ICU patients, with the assumption that the user will subselect appropriate ICUSTAY_IDs.
+-- For example, the score is calculated for neonates, but it is likely inappropriate to actually use the score values for these patients.
+
+with bg as
+(
+ -- join blood gas to ventilation durations to determine if patient was vent
+ select bg.icustay_id
+ , min(pco2) as paco2_min
+ from blood_gas_first_day_arterial bg
+ where specimen_pred = 'ART'
+ group by bg.icustay_id
+)
+-- Aggregate the components for the score
+, scorecomp as
+(
+select ie.icustay_id
+ , v.tempc_min
+ , v.tempc_max
+ , v.heartrate_max
+ , v.resprate_max
+ , bg.paco2_min
+ , l.wbc_min
+ , l.wbc_max
+ , l.bands_max
+from icustays ie
+left join bg
+ on ie.icustay_id = bg.icustay_id
+left join vitals_first_day v
+ on ie.icustay_id = v.icustay_id
+left join labs_first_day l
+ on ie.icustay_id = l.icustay_id
+)
+, scorecalc as
+(
+ -- Calculate the final score
+ -- note that if the underlying data is missing, the component is null
+ -- eventually these are treated as 0 (normal), but knowing when data is missing is useful for debugging
+ select icustay_id
+
+ , case
+ when tempc_min < 36.0 then 1
+ when tempc_max > 38.0 then 1
+ when tempc_min is null then null
+ else 0
+ end as temp_score
+
+
+ , case
+ when heartrate_max > 90.0 then 1
+ when heartrate_max is null then null
+ else 0
+ end as heartrate_score
+
+ , case
+ when resprate_max > 20.0 then 1
+ when paco2_min < 32.0 then 1
+ when coalesce(resprate_max, paco2_min) is null then null
+ else 0
+ end as resp_score
+
+ , case
+ when wbc_min < 4.0 then 1
+ when wbc_max > 12.0 then 1
+ when bands_max > 10 then 1-- > 10% immature neurophils (band forms)
+ when coalesce(wbc_min, bands_max) is null then null
+ else 0
+ end as wbc_score
+
+ from scorecomp
+)
+select
+ ie.subject_id, ie.hadm_id, ie.icustay_id
+ -- Combine all the scores to get SOFA
+ -- Impute 0 if the score is missing
+ , coalesce(temp_score,0)
+ + coalesce(heartrate_score,0)
+ + coalesce(resp_score,0)
+ + coalesce(wbc_score,0)
+ as sirs
+ , temp_score, heartrate_score, resp_score, wbc_score
+from icustays ie
+left join scorecalc s
+ on ie.icustay_id = s.icustay_id
+order by ie.icustay_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/49_qsofa.sql b/mimic-iii-carevue/code/concepts/49_qsofa.sql
new file mode 100644
index 000000000..c0a044ff2
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/49_qsofa.sql
@@ -0,0 +1,70 @@
+drop table if exists qsofa; create table qsofa as
+-- ------------------------------------------------------------------
+-- Title: Quick Sequential Organ Failure Assessment (qSOFA)
+-- This query extracts the quick sequential organ failure assessment.
+-- This score was a recent revision of SOFA, aiming to detect patients at risk of sepsis.
+-- The score is calculated on the first day of each ICU patients' stay - though needn't be.
+-- ------------------------------------------------------------------
+
+-- Reference for qSOFA:
+-- Singer M, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3)
+-- Seymour CW, et al. Assessment of Clinical Criteria for Sepsis: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3)
+
+-- Variables used in qSOFA:
+-- GCS, respiratory rate, systolic blood pressure
+
+-- The following views required to run this query:
+-- 1) gcsfirstday - generated by gcs-first-day.sql
+-- 2) vitalsfirstday - generated by vitals-first-day.sql
+
+-- Note:
+-- The score is calculated for *all* ICU patients, with the assumption that the user will subselect appropriate ICUSTAY_IDs.
+-- For example, the score is calculated for neonates, but it is likely inappropriate to actually use the score values for these patients.
+
+with scorecomp as
+(
+select ie.icustay_id
+ , v.sysbp_min
+ , v.resprate_max
+ , gcs.mingcs
+from icustays ie
+left join vitals_first_day v
+ on ie.icustay_id = v.icustay_id
+left join gcs_first_day gcs
+ on ie.icustay_id = gcs.icustay_id
+)
+, scorecalc as
+(
+ -- Calculate the final score
+ -- note that if the underlying data is missing, the component is null
+ -- eventually these are treated as 0 (normal), but knowing when data is missing is useful for debugging
+ select icustay_id
+ , case
+ when sysbp_min is null then null
+ when sysbp_min <= 100 then 1
+ else 0 end
+ as sysbp_score
+ , case
+ when mingcs is null then null
+ when mingcs <= 13 then 1
+ else 0 end
+ as gcs_score
+ , case
+ when resprate_max is null then null
+ when resprate_max >= 22 then 1
+ else 0 end
+ as resprate_score
+ from scorecomp
+)
+select ie.subject_id, ie.hadm_id, ie.icustay_id
+, coalesce(sysbp_score,0)
+ + coalesce(gcs_score,0)
+ + coalesce(resprate_score,0)
+ as qsofa
+, sysbp_score
+, gcs_score
+, resprate_score
+from icustays ie
+left join scorecalc s
+ on ie.icustay_id = s.icustay_id
+order by ie.icustay_id;
diff --git a/mimic-iii-carevue/code/concepts/50_sepsis3.sql b/mimic-iii-carevue/code/concepts/50_sepsis3.sql
new file mode 100644
index 000000000..0ca9afa07
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/50_sepsis3.sql
@@ -0,0 +1,375 @@
+-- https://github.com/alistairewj/sepsis3-mimic
+-- Johnson AEW, Aboab J, Raffa JD, Pollard TJ, Deliberato RO, Celi LA, Stone DJ. A Comparative Analysis of Sepsis Identification Methods in an Electronic Database. Crit Care Med. 2018 Apr;46(4):494-499. doi: 10.1097/CCM.0000000000002965. PMID: 29303796; PMCID: PMC5851804.
+
+drop table if exists sepsis3; create table sepsis3 as
+-- abx_poe_list
+with t1 as (
+ select
+ drug, drug_name_generic
+ , route
+ , case
+ when lower(drug) like '%' || lower('adoxa') || '%' then 1
+ when lower(drug) like '%' || lower('ala-tet') || '%' then 1
+ when lower(drug) like '%' || lower('alodox') || '%' then 1
+ when lower(drug) like '%' || lower('amikacin') || '%' then 1
+ when lower(drug) like '%' || lower('amikin') || '%' then 1
+ when lower(drug) like '%' || lower('amoxicillin') || '%' then 1
+ when lower(drug) like '%' || lower('amoxicillin%clavulanate') || '%' then 1
+ when lower(drug) like '%' || lower('clavulanate') || '%' then 1
+ when lower(drug) like '%' || lower('ampicillin') || '%' then 1
+ when lower(drug) like '%' || lower('augmentin') || '%' then 1
+ when lower(drug) like '%' || lower('avelox') || '%' then 1
+ when lower(drug) like '%' || lower('avidoxy') || '%' then 1
+ when lower(drug) like '%' || lower('azactam') || '%' then 1
+ when lower(drug) like '%' || lower('azithromycin') || '%' then 1
+ when lower(drug) like '%' || lower('aztreonam') || '%' then 1
+ when lower(drug) like '%' || lower('axetil') || '%' then 1
+ when lower(drug) like '%' || lower('bactocill') || '%' then 1
+ when lower(drug) like '%' || lower('bactrim') || '%' then 1
+ when lower(drug) like '%' || lower('bethkis') || '%' then 1
+ when lower(drug) like '%' || lower('biaxin') || '%' then 1
+ when lower(drug) like '%' || lower('bicillin l-a') || '%' then 1
+ when lower(drug) like '%' || lower('cayston') || '%' then 1
+ when lower(drug) like '%' || lower('cefazolin') || '%' then 1
+ when lower(drug) like '%' || lower('cedax') || '%' then 1
+ when lower(drug) like '%' || lower('cefoxitin') || '%' then 1
+ when lower(drug) like '%' || lower('ceftazidime') || '%' then 1
+ when lower(drug) like '%' || lower('cefaclor') || '%' then 1
+ when lower(drug) like '%' || lower('cefadroxil') || '%' then 1
+ when lower(drug) like '%' || lower('cefdinir') || '%' then 1
+ when lower(drug) like '%' || lower('cefditoren') || '%' then 1
+ when lower(drug) like '%' || lower('cefepime') || '%' then 1
+ when lower(drug) like '%' || lower('cefotetan') || '%' then 1
+ when lower(drug) like '%' || lower('cefotaxime') || '%' then 1
+ when lower(drug) like '%' || lower('cefpodoxime') || '%' then 1
+ when lower(drug) like '%' || lower('cefprozil') || '%' then 1
+ when lower(drug) like '%' || lower('ceftibuten') || '%' then 1
+ when lower(drug) like '%' || lower('ceftin') || '%' then 1
+ when lower(drug) like '%' || lower('cefuroxime ') || '%' then 1
+ when lower(drug) like '%' || lower('cefuroxime') || '%' then 1
+ when lower(drug) like '%' || lower('cephalexin') || '%' then 1
+ when lower(drug) like '%' || lower('chloramphenicol') || '%' then 1
+ when lower(drug) like '%' || lower('cipro') || '%' then 1
+ when lower(drug) like '%' || lower('ciprofloxacin') || '%' then 1
+ when lower(drug) like '%' || lower('claforan') || '%' then 1
+ when lower(drug) like '%' || lower('clarithromycin') || '%' then 1
+ when lower(drug) like '%' || lower('cleocin') || '%' then 1
+ when lower(drug) like '%' || lower('clindamycin') || '%' then 1
+ when lower(drug) like '%' || lower('cubicin') || '%' then 1
+ when lower(drug) like '%' || lower('dicloxacillin') || '%' then 1
+ when lower(drug) like '%' || lower('doryx') || '%' then 1
+ when lower(drug) like '%' || lower('doxycycline') || '%' then 1
+ when lower(drug) like '%' || lower('duricef') || '%' then 1
+ when lower(drug) like '%' || lower('dynacin') || '%' then 1
+ when lower(drug) like '%' || lower('ery-tab') || '%' then 1
+ when lower(drug) like '%' || lower('eryped') || '%' then 1
+ when lower(drug) like '%' || lower('eryc') || '%' then 1
+ when lower(drug) like '%' || lower('erythrocin') || '%' then 1
+ when lower(drug) like '%' || lower('erythromycin') || '%' then 1
+ when lower(drug) like '%' || lower('factive') || '%' then 1
+ when lower(drug) like '%' || lower('flagyl') || '%' then 1
+ when lower(drug) like '%' || lower('fortaz') || '%' then 1
+ when lower(drug) like '%' || lower('furadantin') || '%' then 1
+ when lower(drug) like '%' || lower('garamycin') || '%' then 1
+ when lower(drug) like '%' || lower('gentamicin') || '%' then 1
+ when lower(drug) like '%' || lower('kanamycin') || '%' then 1
+ when lower(drug) like '%' || lower('keflex') || '%' then 1
+ when lower(drug) like '%' || lower('ketek') || '%' then 1
+ when lower(drug) like '%' || lower('levaquin') || '%' then 1
+ when lower(drug) like '%' || lower('levofloxacin') || '%' then 1
+ when lower(drug) like '%' || lower('lincocin') || '%' then 1
+ when lower(drug) like '%' || lower('macrobid') || '%' then 1
+ when lower(drug) like '%' || lower('macrodantin') || '%' then 1
+ when lower(drug) like '%' || lower('maxipime') || '%' then 1
+ when lower(drug) like '%' || lower('mefoxin') || '%' then 1
+ when lower(drug) like '%' || lower('metronidazole') || '%' then 1
+ when lower(drug) like '%' || lower('minocin') || '%' then 1
+ when lower(drug) like '%' || lower('minocycline') || '%' then 1
+ when lower(drug) like '%' || lower('monodox') || '%' then 1
+ when lower(drug) like '%' || lower('monurol') || '%' then 1
+ when lower(drug) like '%' || lower('morgidox') || '%' then 1
+ when lower(drug) like '%' || lower('moxatag') || '%' then 1
+ when lower(drug) like '%' || lower('moxifloxacin') || '%' then 1
+ when lower(drug) like '%' || lower('myrac') || '%' then 1
+ when lower(drug) like '%' || lower('nafcillin sodium') || '%' then 1
+ when lower(drug) like '%' || lower('nicazel doxy 30') || '%' then 1
+ when lower(drug) like '%' || lower('nitrofurantoin') || '%' then 1
+ when lower(drug) like '%' || lower('noroxin') || '%' then 1
+ when lower(drug) like '%' || lower('ocudox') || '%' then 1
+ when lower(drug) like '%' || lower('ofloxacin') || '%' then 1
+ when lower(drug) like '%' || lower('omnicef') || '%' then 1
+ when lower(drug) like '%' || lower('oracea') || '%' then 1
+ when lower(drug) like '%' || lower('oraxyl') || '%' then 1
+ when lower(drug) like '%' || lower('oxacillin') || '%' then 1
+ when lower(drug) like '%' || lower('pc pen vk') || '%' then 1
+ when lower(drug) like '%' || lower('pce dispertab') || '%' then 1
+ when lower(drug) like '%' || lower('panixine') || '%' then 1
+ when lower(drug) like '%' || lower('pediazole') || '%' then 1
+ when lower(drug) like '%' || lower('penicillin') || '%' then 1
+ when lower(drug) like '%' || lower('periostat') || '%' then 1
+ when lower(drug) like '%' || lower('pfizerpen') || '%' then 1
+ when lower(drug) like '%' || lower('piperacillin') || '%' then 1
+ when lower(drug) like '%' || lower('tazobactam') || '%' then 1
+ when lower(drug) like '%' || lower('primsol') || '%' then 1
+ when lower(drug) like '%' || lower('proquin') || '%' then 1
+ when lower(drug) like '%' || lower('raniclor') || '%' then 1
+ when lower(drug) like '%' || lower('rifadin') || '%' then 1
+ when lower(drug) like '%' || lower('rifampin') || '%' then 1
+ when lower(drug) like '%' || lower('rocephin') || '%' then 1
+ when lower(drug) like '%' || lower('smz-tmp') || '%' then 1
+ when lower(drug) like '%' || lower('septra') || '%' then 1
+ when lower(drug) like '%' || lower('septra ds') || '%' then 1
+ when lower(drug) like '%' || lower('septra') || '%' then 1
+ when lower(drug) like '%' || lower('solodyn') || '%' then 1
+ when lower(drug) like '%' || lower('spectracef') || '%' then 1
+ when lower(drug) like '%' || lower('streptomycin sulfate') || '%' then 1
+ when lower(drug) like '%' || lower('sulfadiazine') || '%' then 1
+ when lower(drug) like '%' || lower('sulfamethoxazole') || '%' then 1
+ when lower(drug) like '%' || lower('trimethoprim') || '%' then 1
+ when lower(drug) like '%' || lower('sulfatrim') || '%' then 1
+ when lower(drug) like '%' || lower('sulfisoxazole') || '%' then 1
+ when lower(drug) like '%' || lower('suprax') || '%' then 1
+ when lower(drug) like '%' || lower('synercid') || '%' then 1
+ when lower(drug) like '%' || lower('tazicef') || '%' then 1
+ when lower(drug) like '%' || lower('tetracycline') || '%' then 1
+ when lower(drug) like '%' || lower('timentin') || '%' then 1
+ when lower(drug) like '%' || lower('tobi') || '%' then 1
+ when lower(drug) like '%' || lower('tobramycin') || '%' then 1
+ when lower(drug) like '%' || lower('trimethoprim') || '%' then 1
+ when lower(drug) like '%' || lower('unasyn') || '%' then 1
+ when lower(drug) like '%' || lower('vancocin') || '%' then 1
+ when lower(drug) like '%' || lower('vancomycin') || '%' then 1
+ when lower(drug) like '%' || lower('vantin') || '%' then 1
+ when lower(drug) like '%' || lower('vibativ') || '%' then 1
+ when lower(drug) like '%' || lower('vibra-tabs') || '%' then 1
+ when lower(drug) like '%' || lower('vibramycin') || '%' then 1
+ when lower(drug) like '%' || lower('zinacef') || '%' then 1
+ when lower(drug) like '%' || lower('zithromax') || '%' then 1
+ when lower(drug) like '%' || lower('zmax') || '%' then 1
+ when lower(drug) like '%' || lower('zosyn') || '%' then 1
+ when lower(drug) like '%' || lower('zyvox') || '%' then 1
+ else 0
+ end as antibiotic
+ from prescriptions
+ where drug_type in ('MAIN','ADDITIVE')
+ -- we exclude routes via the eye, ears, or topically
+ and route not in ('OU','OS','OD','AU','AS','AD', 'TP')
+ and route not ilike '%ear%'
+ and route not ilike '%eye%'
+ -- we exclude certain types of antibiotics: topical creams, gels, desens, etc
+ and drug not ilike '%cream%'
+ and drug not ilike '%desensitization%'
+ and drug not ilike '%ophth oint%'
+ and drug not ilike '%gel%'
+ -- other routes not sure about...
+ -- for sure keep: ('IV','PO','PO/NG','ORAL', 'IV DRIP', 'IV BOLUS')
+ -- ? VT, PB, PR, PL, NS, NG, NEB, NAS, LOCK, J TUBE, IVT
+ -- ? IT, IRR, IP, IO, INHALATION, IN, IM
+ -- ? IJ, IH, G TUBE, DIALYS
+ -- ?? enemas??
+)
+
+, abx_poe_list as (
+ select
+ drug --, drug_name_generic
+ , count(*) as numobs
+ from t1
+ where antibiotic = 1
+ group by drug --, drug_name_generic
+ order by numobs desc
+)
+
+
+
+-- abx_micro_poe
+-- defines suspicion of infection using prescriptions + microbiologyevents
+, abx as (
+ select pr.hadm_id
+ , pr.drug as antibiotic_name
+ , pr.startdate as antibiotic_time
+ , pr.enddate as antibiotic_endtime
+ from prescriptions pr
+ -- inner join to subselect to only antibiotic prescriptions
+ inner join abx_poe_list ab
+ on pr.drug = ab.drug
+)
+-- get cultures for each icustay
+-- note this duplicates prescriptions
+-- each ICU stay in the same hospitalization will get a copy of all prescriptions for that hospitalization
+, ab_tbl as (
+ select
+ ie.subject_id, ie.hadm_id, ie.icustay_id
+ , ie.intime, ie.outtime
+ , abx.antibiotic_name
+ , abx.antibiotic_time
+ , abx.antibiotic_endtime
+ from icustays ie
+ left join abx
+ on ie.hadm_id = abx.hadm_id
+)
+, me as (
+ select hadm_id
+ , chartdate, charttime
+ , spec_type_desc
+ , max(case when org_name is not null and org_name != '' then 1 else 0 end) as PositiveCulture
+ from microbiologyevents
+ group by hadm_id, chartdate, charttime, spec_type_desc
+)
+, ab_fnl as (
+ select
+ ab_tbl.icustay_id, ab_tbl.intime, ab_tbl.outtime
+ , ab_tbl.antibiotic_name
+ , ab_tbl.antibiotic_time
+ , coalesce(me72.charttime,me72.chartdate) as last72_charttime
+ , coalesce(me24.charttime,me24.chartdate) as next24_charttime
+
+ , me72.positiveculture as last72_positiveculture
+ , me72.spec_type_desc as last72_specimen
+ , me24.positiveculture as next24_positiveculture
+ , me24.spec_type_desc as next24_specimen
+ from ab_tbl
+ -- blood culture in last 72 hours
+ left join me me72
+ on ab_tbl.hadm_id = me72.hadm_id
+ and ab_tbl.antibiotic_time is not null
+ and
+ (
+ -- if charttime is available, use it
+ (
+ ab_tbl.antibiotic_time > me72.charttime
+ and ab_tbl.antibiotic_time <= me72.charttime + interval '72' hour
+ )
+ or
+ (
+ -- if charttime is not available, use chartdate
+ me72.charttime is null
+ and ab_tbl.antibiotic_time > me72.chartdate
+ and ab_tbl.antibiotic_time < me72.chartdate + interval '96' hour -- could equally do this with a date_trunc, but that's less portable
+ )
+ )
+ -- blood culture in subsequent 24 hours
+ left join me me24
+ on ab_tbl.hadm_id = me24.hadm_id
+ and ab_tbl.antibiotic_time is not null
+ and me24.charttime is not null
+ and
+ (
+ -- if charttime is available, use it
+ (
+ ab_tbl.antibiotic_time > me24.charttime - interval '24' hour
+ and ab_tbl.antibiotic_time <= me24.charttime
+ )
+ or
+ (
+ -- if charttime is not available, use chartdate
+ me24.charttime is null
+ and ab_tbl.antibiotic_time > me24.chartdate
+ and ab_tbl.antibiotic_time <= me24.chartdate + interval '24' hour
+ )
+ )
+)
+, ab_laststg as (
+select
+ icustay_id
+ , antibiotic_name
+ , antibiotic_time
+ , last72_charttime
+ , next24_charttime
+
+ -- time of suspected infection: either the culture time (if before antibiotic), or the antibiotic time
+ , case
+ when coalesce(last72_charttime,antibiotic_time) is null
+ then 0
+ else 1 end as suspected_infection
+
+ , coalesce(last72_charttime,antibiotic_time) as suspected_infection_time
+
+ -- the specimen that was cultured
+ , case
+ when last72_charttime is not null
+ then last72_specimen
+ when next24_charttime is not null
+ then next24_specimen
+ else null
+ end as specimen
+
+ -- whether the cultured specimen ended up being positive or not
+ , case
+ when last72_charttime is not null
+ then last72_positiveculture
+ when next24_charttime is not null
+ then next24_positiveculture
+ else null
+ end as positiveculture
+from ab_fnl
+)
+
+, abx_micro_poe as (
+ select
+ icustay_id
+ , antibiotic_name
+ , antibiotic_time
+ , last72_charttime
+ , next24_charttime
+ , suspected_infection_time
+ -- -- the below two fields are used to extract data - modifying them facilitates sensitivity analyses
+ -- , suspected_infection_time - interval '48' hour as si_starttime
+ -- , suspected_infection_time + interval '24' hour as si_endtime
+ , specimen, positiveculture
+ from ab_laststg
+ order by icustay_id, antibiotic_time
+)
+
+-- suspinfect_poe
+, susp_abx as
+(
+ select icustay_id
+ , suspected_infection_time
+ , specimen, positiveculture
+ , antibiotic_name
+ , antibiotic_time
+ , row_number() over (partition by icustay_id order by suspected_infection_time) as rn
+ from abx_micro_poe
+)
+
+, suspinfect_poe as (
+ select
+ ie.icustay_id
+ , ie.intime
+ , antibiotic_name
+ , antibiotic_time
+ , suspected_infection_time as suspected_infection_time_poe
+ , extract(EPOCH from ie.intime - suspected_infection_time) / 60.0 / 60.0 / 24.0 as suspected_infection_time_poe_days
+ , specimen
+ , positiveculture
+ from icustays ie
+ left join susp_abx sa
+ on ie.icustay_id = sa.icustay_id
+ and sa.rn = 1
+ order by ie.icustay_id
+)
+
+, sepsis3_info as (
+ select
+ sp.icustay_id,
+ sofa,
+ case
+ when suspected_infection_time_poe_days is not null then 1
+ else 0
+ end as suspicion_poe
+ from
+ suspinfect_poe sp
+ left join sofa so
+ using(icustay_id)
+)
+
+select
+ icustay_id,
+ case
+ when sofa >= 2 and suspicion_poe = 1 then 1
+ else 0
+ end as sepsis3
+from
+ sepsis3_info
+order by
+ icustay_id;
diff --git a/mimic-iii-carevue/code/concepts/51_age.sql b/mimic-iii-carevue/code/concepts/51_age.sql
new file mode 100644
index 000000000..866f622d9
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/51_age.sql
@@ -0,0 +1,29 @@
+
+-- THIS SCRIPT IS AUTOMATICALLY GENERATED. DO NOT EDIT IT DIRECTLY.
+drop table if exists age; create table age as
+
+-- https://mimic.mit.edu/docs/iii/about/time/#dates-of-birth
+
+-- Dates of birth which occur in the present time are not true dates of birth.
+-- Furthermore, dates of birth which occur before the year 1900 occur if the patient is older than 89.
+-- In these cases, the patient’s age at their first admission has been fixed to 300.
+
+with age_raw as (
+ select
+ icustay_id,
+ hadm_id,
+ round(cast(extract(epoch from (admittime - dob))/(60*60*24*365) as numeric), 2) as age
+ from
+ mimiciii_cv.icustays
+ left join mimiciii_cv.patients
+ using (subject_id)
+ left join mimiciii_cv.admissions
+ using (hadm_id)
+)
+
+select
+ distinct
+ hadm_id,
+ case when age >= 90 then 90 else age end as age
+from
+ age_raw;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/52_charlson.sql b/mimic-iii-carevue/code/concepts/52_charlson.sql
new file mode 100644
index 000000000..1a2b9f05d
--- /dev/null
+++ b/mimic-iii-carevue/code/concepts/52_charlson.sql
@@ -0,0 +1,226 @@
+-- THIS SCRIPT IS AUTOMATICALLY GENERATED. DO NOT EDIT IT DIRECTLY.
+drop table if exists charlson; create table charlson as
+-- ------------------------------------------------------------------
+-- This query extracts Charlson Comorbidity Index (CCI) based on the recorded ICD-9 codes.
+--
+-- Reference for CCI:
+-- (1) Charlson ME, Pompei P, Ales KL, MacKenzie CR. (1987) A new method of classifying prognostic
+-- comorbidity in longitudinal studies: development and validation.J Chronic Dis; 40(5):373-83.
+--
+-- (2) Charlson M, Szatrowski TP, Peterson J, Gold J. (1994) Validation of a combined comorbidity
+-- index. J Clin Epidemiol; 47(11):1245-51.
+--
+-- Reference for ICD-9-CM Coding Algorithm for Charlson Comorbidities:
+-- (3) Quan H, Sundararajan V, Halfon P, et al. Coding algorithms for defining Comorbidities in ICD-9-CM
+-- and ICD-10 administrative data. Med Care. 2005 Nov; 43(11): 1130-9.
+-- ------------------------------------------------------------------
+
+with diag as (
+ select
+ hadm_id,
+ icd9_code
+ from
+ diagnoses_icd
+)
+
+, com as (
+ select
+ ad.hadm_id
+
+ -- Myocardial infarction
+ , max(case when
+ substr(icd9_code, 1, 3) in ('410','412')
+ then 1
+ else 0 end) as myocardial_infarct
+
+ -- Congestive heart failure
+ , max(case when
+ substr(icd9_code, 1, 3) = '428'
+ or
+ substr(icd9_code, 1, 5) in ('39891','40201','40211','40291','40401','40403','40411','40413','40491','40493')
+ or
+ substr(icd9_code, 1, 4) between '4254' and '4259'
+ then 1
+ else 0 end) as congestive_heart_failure
+
+ -- Peripheral vascular disease
+ , max(case when
+ substr(icd9_code, 1, 3) in ('440','441')
+ or
+ substr(icd9_code, 1, 4) in ('0930','4373','4471','5571','5579','V434')
+ or
+ substr(icd9_code, 1, 4) between '4431' and '4439'
+ then 1
+ else 0 end) as peripheral_vascular_disease
+
+ -- Cerebrovascular disease
+ , max(case when
+ substr(icd9_code, 1, 3) between '430' and '438'
+ or
+ substr(icd9_code, 1, 5) = '36234'
+ then 1
+ else 0 end) as cerebrovascular_disease
+
+ -- Dementia
+ , max(case when
+ substr(icd9_code, 1, 3) = '290'
+ or
+ substr(icd9_code, 1, 4) in ('2941','3312')
+ then 1
+ else 0 end) as dementia
+
+ -- Chronic pulmonary disease
+ , max(case when
+ substr(icd9_code, 1, 3) between '490' and '505'
+ or
+ substr(icd9_code, 1, 4) in ('4168','4169','5064','5081','5088')
+ then 1
+ else 0 end) as chronic_pulmonary_disease
+
+ -- Rheumatic disease
+ , max(case when
+ substr(icd9_code, 1, 3) = '725'
+ or
+ substr(icd9_code, 1, 4) in ('4465','7100','7101','7102','7103','7104','7140','7141','7142','7148')
+ then 1
+ else 0 end) as rheumatic_disease
+
+ -- Peptic ulcer disease
+ , max(case when
+ substr(icd9_code, 1, 3) in ('531','532','533','534')
+ then 1
+ else 0 end) as peptic_ulcer_disease
+
+ -- Mild liver disease
+ , max(case when
+ substr(icd9_code, 1, 3) in ('570','571')
+ or
+ substr(icd9_code, 1, 4) in ('0706','0709','5733','5734','5738','5739','V427')
+ or
+ substr(icd9_code, 1, 5) in ('07022','07023','07032','07033','07044','07054')
+ then 1
+ else 0 end) as mild_liver_disease
+
+ -- Diabetes without chronic complication
+ , max(case when
+ substr(icd9_code, 1, 4) in ('2500','2501','2502','2503','2508','2509')
+ then 1
+ else 0 end) as diabetes_without_cc
+
+ -- Diabetes with chronic complication
+ , max(case when
+ substr(icd9_code, 1, 4) in ('2504','2505','2506','2507')
+ then 1
+ else 0 end) as diabetes_with_cc
+
+ -- Hemiplegia or paraplegia
+ , max(case when
+ substr(icd9_code, 1, 3) in ('342','343')
+ or
+ substr(icd9_code, 1, 4) in ('3341','3440','3441','3442','3443','3444','3445','3446','3449')
+ then 1
+ else 0 end) as paraplegia
+
+ -- Renal disease
+ , max(case when
+ substr(icd9_code, 1, 3) in ('582','585','586','V56')
+ or
+ substr(icd9_code, 1, 4) in ('5880','V420','V451')
+ or
+ substr(icd9_code, 1, 4) between '5830' and '5837'
+ or
+ substr(icd9_code, 1, 5) in ('40301','40311','40391','40402','40403','40412','40413','40492','40493')
+ then 1
+ else 0 end) as renal_disease
+
+ -- Any malignancy, including lymphoma and leukemia, except malignant neoplasm of skin
+ , max(case when
+ substr(icd9_code, 1, 3) between '140' and '172'
+ or
+ substr(icd9_code, 1, 4) between '1740' and '1958'
+ or
+ substr(icd9_code, 1, 3) between '200' and '208'
+ or
+ substr(icd9_code, 1, 4) = '2386'
+ then 1
+ else 0 end) as malignant_cancer
+
+ -- Moderate or severe liver disease
+ , max(case when
+ substr(icd9_code, 1, 4) in ('4560','4561','4562')
+ or
+ substr(icd9_code, 1, 4) between '5722' and '5728'
+ then 1
+ else 0 end) as severe_liver_disease
+
+ -- Metastatic solid tumor
+ , max(case when
+ substr(icd9_code, 1, 3) in ('196','197','198','199')
+ then 1
+ else 0 end) as metastatic_solid_tumor
+
+ -- AIDS/HIV
+ , max(case when
+ substr(icd9_code, 1, 3) in ('042','043','044')
+ then 1
+ else 0 end) as aids
+ from
+ admissions ad
+ left join diag
+ on ad.hadm_id = diag.hadm_id
+ group by ad.hadm_id
+)
+
+, ag as (
+ select
+ hadm_id,
+ age,
+ case when age <= 40 then 0
+ when age <= 50 then 1
+ when age <= 60 then 2
+ when age <= 70 then 3
+ else 4
+ end as age_score
+ from
+ age
+)
+
+select
+ ad.subject_id,
+ ad.hadm_id,
+ ag.age_score,
+ myocardial_infarct,
+ congestive_heart_failure,
+ peripheral_vascular_disease,
+ cerebrovascular_disease,
+ dementia,
+ chronic_pulmonary_disease,
+ rheumatic_disease,
+ peptic_ulcer_disease,
+ mild_liver_disease,
+ diabetes_without_cc,
+ diabetes_with_cc,
+ paraplegia,
+ renal_disease,
+ malignant_cancer,
+ severe_liver_disease,
+ metastatic_solid_tumor,
+ aids,
+ -- Calculate the Charlson Comorbidity Score using the original
+ -- weights from Charlson, 1987.
+ age_score
+ + myocardial_infarct + congestive_heart_failure + peripheral_vascular_disease
+ + cerebrovascular_disease + dementia + chronic_pulmonary_disease
+ + rheumatic_disease + peptic_ulcer_disease
+ + greatest(mild_liver_disease, 3*severe_liver_disease)
+ + greatest(2*diabetes_with_cc, diabetes_without_cc)
+ + greatest(2*malignant_cancer, 6*metastatic_solid_tumor)
+ + 2*paraplegia + 2*renal_disease
+ + 6*aids
+ as charlson_comorbidity_index
+from
+ admissions ad
+left join com
+ on ad.hadm_id = com.hadm_id
+left join ag
+ on com.hadm_id = ag.hadm_id;
\ No newline at end of file
diff --git a/mimic-iii-carevue/code/concepts/dx_data/ccs_multi_dx.csv.gz b/mimic-iii-carevue/code/concepts/dx_data/ccs_multi_dx.csv.gz
new file mode 100644
index 000000000..5e13857a6
Binary files /dev/null and b/mimic-iii-carevue/code/concepts/dx_data/ccs_multi_dx.csv.gz differ
diff --git a/mimic-iii-carevue/img/01.png b/mimic-iii-carevue/img/01.png
new file mode 100644
index 000000000..457debd51
Binary files /dev/null and b/mimic-iii-carevue/img/01.png differ
diff --git a/mimic-iii-carevue/img/02.png b/mimic-iii-carevue/img/02.png
new file mode 100644
index 000000000..a4a0c0013
Binary files /dev/null and b/mimic-iii-carevue/img/02.png differ
diff --git a/mimic-iii-carevue/img/03.png b/mimic-iii-carevue/img/03.png
new file mode 100644
index 000000000..db7a18b2d
Binary files /dev/null and b/mimic-iii-carevue/img/03.png differ
diff --git a/mimic-iii-carevue/img/04.png b/mimic-iii-carevue/img/04.png
new file mode 100644
index 000000000..ec2bd2da4
Binary files /dev/null and b/mimic-iii-carevue/img/04.png differ
diff --git a/mimic-iii-carevue/img/05.png b/mimic-iii-carevue/img/05.png
new file mode 100644
index 000000000..f6961142f
Binary files /dev/null and b/mimic-iii-carevue/img/05.png differ
diff --git a/mimic-iii-carevue/img/06.png b/mimic-iii-carevue/img/06.png
new file mode 100644
index 000000000..0c80994bc
Binary files /dev/null and b/mimic-iii-carevue/img/06.png differ
diff --git a/mimic-iii-carevue/img/07.png b/mimic-iii-carevue/img/07.png
new file mode 100644
index 000000000..fb469defa
Binary files /dev/null and b/mimic-iii-carevue/img/07.png differ
diff --git a/mimic-iii-carevue/img/08.png b/mimic-iii-carevue/img/08.png
new file mode 100644
index 000000000..46b6b02c3
Binary files /dev/null and b/mimic-iii-carevue/img/08.png differ
diff --git a/mimic-iii-carevue/img/09.png b/mimic-iii-carevue/img/09.png
new file mode 100644
index 000000000..db0c5e2d9
Binary files /dev/null and b/mimic-iii-carevue/img/09.png differ
diff --git a/mimic-iii-carevue/img/10.png b/mimic-iii-carevue/img/10.png
new file mode 100644
index 000000000..979280880
Binary files /dev/null and b/mimic-iii-carevue/img/10.png differ
diff --git a/mimic-iii-carevue/img/11.png b/mimic-iii-carevue/img/11.png
new file mode 100644
index 000000000..52b75dfe2
Binary files /dev/null and b/mimic-iii-carevue/img/11.png differ
diff --git a/mimic-iii-carevue/img/12.png b/mimic-iii-carevue/img/12.png
new file mode 100644
index 000000000..f7712792b
Binary files /dev/null and b/mimic-iii-carevue/img/12.png differ
diff --git a/mimic-iii-carevue/img/13.png b/mimic-iii-carevue/img/13.png
new file mode 100644
index 000000000..18b355dbf
Binary files /dev/null and b/mimic-iii-carevue/img/13.png differ
diff --git a/mimic-iii-carevue/img/14.png b/mimic-iii-carevue/img/14.png
new file mode 100644
index 000000000..277cee84e
Binary files /dev/null and b/mimic-iii-carevue/img/14.png differ
diff --git a/mimic-iii-carevue/img/15.png b/mimic-iii-carevue/img/15.png
new file mode 100644
index 000000000..e29c30d56
Binary files /dev/null and b/mimic-iii-carevue/img/15.png differ
diff --git a/mimic-iii-carevue/install.sh b/mimic-iii-carevue/install.sh
new file mode 100644
index 000000000..fe3400cf5
--- /dev/null
+++ b/mimic-iii-carevue/install.sh
@@ -0,0 +1,578 @@
+#!/bin/bash
+
+db="MIMIC-III CareVue"
+
+echo -e "===================================="
+echo -e "One-Click Installation Universal Script for $db on MAC OS, Windows, and Linux"
+echo -e "===================================="
+echo -e "$db 数据库全平台通用(MAC OS、Windows、Linux)一键安装脚本"
+echo -e "===================================="
+echo -e "ningyile@qq.com"
+echo -e "===================================="
+echo -e "https://github.com/ningyile"
+echo -e "===================================="
+
+
+
+# display language
+while true
+do
+ echo -e "Enter a choice for display language(输入选项以选择语言):
+ 1. English(英文)
+ 2. Chinese(中文)"
+ read lan_sel
+ echo -e "\n"
+ case $lan_sel in
+ 1)
+ echo -e "You select English!\n"
+ ;;
+ 2)
+ echo -e "已选择中文!\n"
+ ;;
+ *)
+ continue
+ esac
+break
+done
+
+
+
+# check whether the current user name meets the requirements
+while true
+do
+ username=$(whoami)
+ # read username
+ if [[ $username =~ ^[a-zA-Z][a-zA-Z0-9_]*$ ]]; then
+ echo -e "\n"
+ else
+ if [ $lan_sel == 1 ]; then
+ # 英文
+ echo -e "The current username does not meet the requirements!"
+ echo -e "The username can contain alphabet(and start with letter), number, and/or underscores(_)!"
+ echo -e "Please abort the installation and add a username that meets the requirements!"
+ echo -e "Are you aware of the requirements and abort the installation?('Y' or 'N')"
+ read abort_sel
+ echo -e "\n"
+ case $abort_sel in
+ 'Y'|'y')
+ # 中止
+ exit
+ ;;
+ *)
+ continue
+ esac
+ else
+ echo -e "当前用户名不符合要求!"
+ echo -e "用户名只能包含英文字母(且字母开头)、数字或者下划线(_)!"
+ echo -e "请中止安装并前往系统添加符合要求的用户名!"
+ echo -e "你已知晓上述要求并中止安装吗?(请输入'Y' 或 'N')"
+ read abort_sel
+ echo -e "\n"
+ case $abort_sel in
+ 'Y'|'y')
+ # 中止
+ exit
+ ;;
+ *)
+ continue
+ esac
+ fi
+ fi
+break
+done
+
+
+
+# reset pg_hba.conf
+user="postgres"
+test_db_login=false
+ptn=" reject$| md5$| password$| scram-sha-256$| gss$| sspi$| ident$| peer$| pam$| ldap$| radius$| cert$"
+sed_ptn="s/$ptn/ trust/g"
+
+if [[ $lan_sel == 1 ]]; then
+ # English
+ while [[ $test_db_login = "false" ]]; do
+
+ unset pass
+ while [ -z $pass ]; do
+ read -sp "Please type the password of database user 'postgres': " pass
+ done
+ echo -e "\n"
+ # set global login password
+ export PGPASSWORD=$pass
+ # get abs path of pg_hba.conf
+ pg_hba=`(psql -U postgres -t -P format=unaligned -c "show hba_file;")`
+
+ if [[ "$pg_hba" =~ "pg_hba.conf" ]]; then
+ test_db_login=true
+ echo "The path of 'pg_hba.conf' is $pg_hba"
+
+ if grep -E "$ptn" "$pg_hba"; then
+ echo -e "===================================="
+ echo -e "\n"
+ echo -e "Please type the password of operating system user!"
+ echo -e "\n"
+ echo -e "===================================="
+ pg_hba_dir=$(sudo dirname "$pg_hba")
+
+ # backup pg_hba.conf
+ cp "$pg_hba" "$pg_hba_dir"/pg_hba.conf.bk
+ # 判断是否是Mac os
+ os="$(uname)"
+ sed_version="$(which sed)"
+
+ # reset the mode of local connection as "trust"
+ if [[ "$os" == "Darwin" ]] && [[ "$sed_version" == "/usr/bin/sed" ]]; then
+
+ sed -i.bk -E "$sed_ptn" "$pg_hba" # FreeBSD-sed
+ else
+ sed -i -E "$sed_ptn" "$pg_hba" # GNU-sed
+ fi
+ echo -e "\n"
+ echo -e "The mode of local connection has been reset as 'trust'!"
+ echo -e "\n"
+ echo -e "===================================="
+ # restart the database to load the configuration
+ psql -U postgres -q -c "SELECT pg_reload_conf();"
+ echo -e "===================================="
+ echo -e "\n"
+ echo -e "Configuration of 'pg_hba.conf' has been reloaded!"
+ echo -e "\n"
+ echo -e "===================================="
+ else
+ echo -e "===================================="
+ echo -e "\n"
+ echo -e "The mode of local connection is 'trust'!"
+ echo -e "\n"
+ echo -e "===================================="
+ fi
+
+ else
+ test_db_login=false
+ echo -e "===================================="
+ echo -e "\n"
+ echo -e "Password authentication failed for user 'postgres'"
+ echo -e "Please retype the correct password of database user 'postgres'!"
+ echo -e "\n"
+ echo -e "===================================="
+ fi
+ done
+
+else
+ # Chinese
+ while [[ $test_db_login = "false" ]]; do
+
+ unset pass
+ while [ -z $pass ]; do
+ read -sp "请输入数据库用户'postgres'的登录密码: " pass
+ done
+ echo -e "\n"
+ # set global login password
+ export PGPASSWORD=$pass
+ # get abs path of pg_hba.conf
+ pg_hba=`(psql -U postgres -t -P format=unaligned -c "show hba_file;")`
+
+ if [[ "$pg_hba" =~ "pg_hba.conf" ]]; then
+ test_db_login=true
+ echo "数据库配置文件'pg_hba.conf'的路径是$pg_hba"
+
+ if grep -E "$ptn" "$pg_hba"; then
+ echo -e "===================================="
+ echo -e "\n"
+ echo -e "请输入操作系统用户登录密码!"
+ echo -e "\n"
+ echo -e "===================================="
+ pg_hba_dir=$(sudo dirname "$pg_hba")
+
+ # backup pg_hba.conf
+ cp "$pg_hba" "$pg_hba_dir"/pg_hba.conf.bk
+ # 判断是否是Mac OS
+ os="$(uname)"
+ sed_version="$(which sed)"
+
+ # reset the mode of local connection as "trust"
+ if [[ "$os" == "Darwin" ]] && [[ "$sed_version" == "/usr/bin/sed" ]]; then
+ sed -i.bk -E "$sed_ptn" "$pg_hba" # FreeBSD-sed
+ else
+ sed -i -E "$sed_ptn" "$pg_hba" # GNU-sed
+ fi
+ echo -e "\n"
+ echo -e "数据库本地连接模式已重置为'trust'!"
+ echo -e "\n"
+ echo -e "===================================="
+ # restart the database to load the configuration
+ psql -U postgres -q -c "SELECT pg_reload_conf();"
+ echo -e "===================================="
+ echo -e "\n"
+ echo -e "数据库配置文件'pg_hba.conf'已重新加载生效!"
+ echo -e "\n"
+ echo -e "===================================="
+ else
+ echo -e "===================================="
+ echo -e "\n"
+ echo -e "数据库本地连接模式为'trust'!"
+ echo -e "\n"
+ echo -e "===================================="
+ fi
+
+ else
+ test_db_login=false
+ echo -e "===================================="
+ echo -e "\n"
+ echo -e "数据库用户'postgres'登录密码输入错误"
+ echo -e "请重新输入数据库用户'postgres'的登录密码!"
+ echo -e "\n"
+ echo -e "===================================="
+ fi
+ done
+fi
+
+
+
+# create database user and database with the same name as the operating system user name
+if [[ $lan_sel == 1 ]]; then
+ # English
+
+ # create database user
+ if psql -U postgres -t -c '\du' | cut -d \| -f 1 | grep -qw "$(whoami)"; then
+ # the database user with the same name as the system user exists
+ echo ""
+ else
+ # the database user with the same name as the system user does not exist
+ echo "--- The database user with the same name as the system user does not exist. ---"
+ echo "--- The database user with the same name as the system user will be created. ---"
+ echo ""
+
+ # create database user with the same name as the system user
+ psql -U postgres -c "create user $(whoami) with superuser createdb createrole replication bypassrls password '$(whoami)';"
+ echo ""
+
+ echo "--- The database user $(whoami) has been created successfully. ---"
+ echo "--- The default password of the database user $(whoami) has been set as::$(whoami) ---"
+ echo ""
+ fi
+
+
+ # create database
+ if psql -U postgres -t -c '\l' | cut -d \| -f 1 | grep -qw "$(whoami)"; then
+ # The database with the same name as the system user exists
+ echo ""
+ else
+ # create database with the same name
+ psql -d postgres -c "create database $(whoami) owner $(whoami);"
+ echo ""
+
+ echo "--- The database with the same name as the system user created successfully. ---"
+ echo "--- You just type 'psql' in the terminal to login Postgresql without any other arguements. ---"
+ echo ""
+ fi
+
+else
+ # Chinese
+
+ # create database user
+ if psql -U postgres -t -c '\du' | cut -d \| -f 1 | grep -qw "$(whoami)"; then
+ # the database user with the same name as the system user exists
+ echo ""
+ else
+ # the database user with the same name as the system user does not exist
+ echo "--- 与系统同名的数据库用户不存在,准备以系统用户为名创建数据库用户 ---"
+ echo ""
+
+ # create database user with the same name as the system user
+ psql -U postgres -c "create user $(whoami) with superuser createdb createrole replication bypassrls password '$(whoami)';"
+ echo ""
+
+ echo "--- 成功创建与系统用户同名的数据库用户$(whoami),用户$(whoami)默认密码已经被设置为:$(whoami) ---"
+ echo ""
+ fi
+
+
+ # create database
+ if psql -U postgres -t -c '\l' | cut -d \| -f 1 | grep -qw "$(whoami)"; then
+ # The database with the same name as the system user exists
+ echo ""
+ else
+ # create database with the same name
+ psql -d postgres -c "create database $(whoami) owner $(whoami);"
+ echo ""
+
+ echo "--- 成功创建与系统用户同名的数据库(方便使用终端登录) ---"
+ echo "--- 你只需在终端输入“psql”即可登录,无需其他任何参数 ---"
+ echo ""
+ fi
+
+fi
+
+
+
+
+# install the database
+if [[ $lan_sel == 1 ]]; then
+ # English
+
+ echo -e "===================================="
+ echo -e 'Any message like "NOTICE: TABLE "XXXXXX" does not exist" during the installation is normal, do not worry about it!'
+ echo -e "===================================="
+ echo ""
+
+ # function of timing
+ secs_to_human() {
+ echo "It takes $(( ${1} / 3600 ))h $(( (${1} / 60) % 60 ))m $(( ${1} % 60 ))s to install the $obj"
+ }
+
+ echo "--- Install basic tables of database ---"
+ echo ""
+ export OP='dbname=mimic3_carevue options=--search_path=mimiciii_cv,mimiciii_derived'
+
+ # build_starttime
+ build_starttime=$(date +%s)
+
+ # drop database if exists
+ psql -c "drop database if exists mimic3_carevue;"
+ # create database
+ psql -c "create database mimic3_carevue owner $(whoami);"
+ # create schema
+ psql -d mimic3_carevue -c "create schema mimiciii_cv;"
+ # create schema
+ psql -d mimic3_carevue -c "create schema mimiciii_derived;"
+ # create tables
+ echo "--- create tables ---"
+ echo ""
+ psql -v on_error_stop=1 -f ./code/buildmimic/01_create_tables.sql "${OP}"
+ # load data
+ echo "--- load data ---"
+ echo ""
+ psql -v on_error_stop=1 -v mimic_data_dir='./database' -f ./code/buildmimic/02_load_data_gz.sql "${OP}"
+ # create indexes
+ echo "--- create indexes ---"
+ echo ""
+ psql -v on_error_stop=1 -f ./code/buildmimic/03_add_indexes.sql "${OP}"
+ # create constraints
+ # Several patients are not in the admission tables, create constraint generates errors, but does not affect.
+ # psql -v on_error_stop=1 -f ./code/buildmimic/04_add_constraints.sql "${OP}"
+
+ echo "--- The basic tables of database has been installed successfully! ---"
+ echo ""
+
+ # build_endtime
+ build_endtime=$(date +%s)
+ # duration of basic tables
+ obj="basic tables of database"
+ secs_to_human "$((${build_endtime} - ${build_starttime}))"
+ echo ""
+
+
+
+ echo "--- Install concepts of database ---"
+ echo ""
+ export OP='dbname=mimic3_carevue options=--search_path=mimiciii_derived,mimiciii_cv'
+
+ # concepts_starttime
+ concepts_starttime=$(date +%s)
+
+ # generate by: ls -a | awk '{print "psql -v on_error_stop=1 -f ./code/concepts/"$0" \"${OP}\""}' > tmp.txt
+ psql -v on_error_stop=1 -f ./code/concepts/01_code_status.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/02_ventilation_classification.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/03_ventilation_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/04_crrt_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/05_dobutamine_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/06_dopamine_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/07_epinephrine_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/08_isuprel_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/09_milrinone_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/10_norepinephrine_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/11_phenylephrine_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/12_vasopressin_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/13_vasopressor_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/14_weight_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/15_elixhauser_ahrq_v37.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/16_elixhauser_ahrq_v37_no_drg.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/17_elixhauser_quan.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/18_elixhauser_score_ahrq.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/19_elixhauser_score_quan.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/20_icustay_detail.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/21_blood_gas_first_day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/22_blood_gas_first_day_arterial.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/23_gcs_first_day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/24_height_first_day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/25_labs_first_day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/26_rrt_first_day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/27_urine_output_first_day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/28_ventilation_first_day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/29_vitals_first_day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/30_weight_first_day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/31_urine_output.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/32_angus.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/33_martin.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/34_explicit.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/35_ccs_dx.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/36_kdigo_creatinine.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/37_kdigo_uo.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/38_kdigo_stages.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/39_kdigo_stages_7day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/40_kdigo_stages_48hr.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/41_meld.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/42_oasis.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/43_sofa.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/44_saps.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/45_sapsii.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/46_apsiii.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/47_lods.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/48_sirs.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/49_qsofa.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/50_sepsis3.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/51_age.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/52_charlson.sql "${OP}"
+
+ echo "--- The concepts of database has been installed successfully! ---"
+ echo ""
+
+ # concepts_endtime
+ concepts_endtime=$(date +%s)
+ # duration of concepts
+ obj="concepts of database"
+ secs_to_human "$((${concepts_endtime} - ${concepts_starttime}))"
+ echo ""
+
+
+
+ # check data
+ echo "--- check data ---"
+ echo ""
+ psql -v on_error_stop=1 -f ./code/buildmimic/05_checks.sql "${OP}"
+
+else
+ # Chinese
+
+ echo -e "===================================="
+ echo -e '期间出现任何类似于 "NOTICE: TABLE "XXXXXX" does not exist"的提示为正常现象,不必多虑!'
+ echo -e "===================================="
+ echo ""
+
+ # 计时函数
+ secs_to_human() {
+ echo "安装 $obj 共耗时$(( ${1} / 3600 ))h $(( (${1} / 60) % 60 ))m $(( ${1} % 60 ))s"
+ }
+
+ echo "--- 安装数据库基础表单 ---"
+ echo ""
+ export OP='dbname=mimic3_carevue options=--search_path=mimiciii_cv,mimiciii_derived'
+
+ # build安装起始时间
+ build_starttime=$(date +%s)
+
+ # drop database if exists
+ psql -c "drop database if exists mimic3_carevue;"
+ # create database
+ psql -c "create database mimic3_carevue owner $(whoami);"
+ # create schema
+ psql -d mimic3_carevue -c "create schema mimiciii_cv;"
+ # create schema
+ psql -d mimic3_carevue -c "create schema mimiciii_derived;"
+ # create tables
+ echo "--- 创建tables ---"
+ echo ""
+ psql -v on_error_stop=1 -f ./code/buildmimic/01_create_tables.sql "${OP}"
+ # load data
+ echo "--- 加载数据 ---"
+ echo ""
+ psql -v on_error_stop=1 -v mimic_data_dir='./database' -f ./code/buildmimic/02_load_data_gz.sql "${OP}"
+ # create indexes
+ echo "--- 创建索引 ---"
+ echo ""
+ psql -v on_error_stop=1 -f ./code/buildmimic/03_add_indexes.sql "${OP}"
+ # create constraints
+ # Several patients are not in the admission tables, create constraint generates errors, but does not affect.
+ # psql -v on_error_stop=1 -f ./code/buildmimic/04_add_constraints.sql "${OP}"
+
+ echo "--- 数据库基础表单安装完成 ---"
+ echo ""
+
+ # build安装结束时间
+ build_endtime=$(date +%s)
+ # 基础表单安装耗时
+ obj="数据库基础表单"
+ secs_to_human "$((${build_endtime} - ${build_starttime}))"
+ echo ""
+
+
+
+ echo "--- 安装数据库Concepts ---"
+ echo ""
+ export OP='dbname=mimic3_carevue options=--search_path=mimiciii_derived,mimiciii_cv'
+
+ # concepts安装起始时间
+ concepts_starttime=$(date +%s)
+
+ # 以下命令由在终端下使用 ls -a | awk '{print "psql -v on_error_stop=1 -f ./code/concepts/"$0" \"${OP}\""}' > tmp.txt 批量生成
+ psql -v on_error_stop=1 -f ./code/concepts/01_code_status.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/02_ventilation_classification.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/03_ventilation_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/04_crrt_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/05_dobutamine_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/06_dopamine_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/07_epinephrine_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/08_isuprel_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/09_milrinone_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/10_norepinephrine_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/11_phenylephrine_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/12_vasopressin_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/13_vasopressor_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/14_weight_durations.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/15_elixhauser_ahrq_v37.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/16_elixhauser_ahrq_v37_no_drg.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/17_elixhauser_quan.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/18_elixhauser_score_ahrq.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/19_elixhauser_score_quan.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/20_icustay_detail.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/21_blood_gas_first_day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/22_blood_gas_first_day_arterial.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/23_gcs_first_day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/24_height_first_day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/25_labs_first_day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/26_rrt_first_day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/27_urine_output_first_day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/28_ventilation_first_day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/29_vitals_first_day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/30_weight_first_day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/31_urine_output.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/32_angus.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/33_martin.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/34_explicit.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/35_ccs_dx.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/36_kdigo_creatinine.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/37_kdigo_uo.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/38_kdigo_stages.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/39_kdigo_stages_7day.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/40_kdigo_stages_48hr.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/41_meld.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/42_oasis.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/43_sofa.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/44_saps.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/45_sapsii.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/46_apsiii.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/47_lods.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/48_sirs.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/49_qsofa.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/50_sepsis3.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/51_age.sql "${OP}"
+ psql -v on_error_stop=1 -f ./code/concepts/52_charlson.sql "${OP}"
+
+ echo "--- 数据库concepts安装完成 ---"
+ echo ""
+
+ # concepts安装结束时间
+ concepts_endtime=$(date +%s)
+ # concepts安装耗时
+ obj="数据库concepts"
+ secs_to_human "$((${concepts_endtime} - ${concepts_starttime}))"
+ echo ""
+
+
+
+ # check data
+ echo "--- 数据核对 ---"
+ echo ""
+ psql -v on_error_stop=1 -f ./code/buildmimic/05_checks.sql "${OP}"
+fi
\ No newline at end of file
 +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+ +
+