@@ -20,6 +19,8 @@
💫 Graph-based foundation model for spatial transcriptomics data
+Novae is a deep learning model for spatial domain assignments of spatial transcriptomics data (at both single-cell or spot resolution). It works across multiple gene panels, tissues, and technologies. Novae offers several additional features, including: (i) native batch-effect correction, (ii) analysis of spatially variable genes and pathways, and (iii) architecture analysis of tissue slides.
+
## Documentation
Check [Novae's documentation](https://mics-lab.github.io/novae/) to get started. It contains installation explanations, API details, and tutorials.
@@ -27,7 +28,7 @@ Check [Novae's documentation](https://mics-lab.github.io/novae/) to get started.
## Overview
-  +
+ 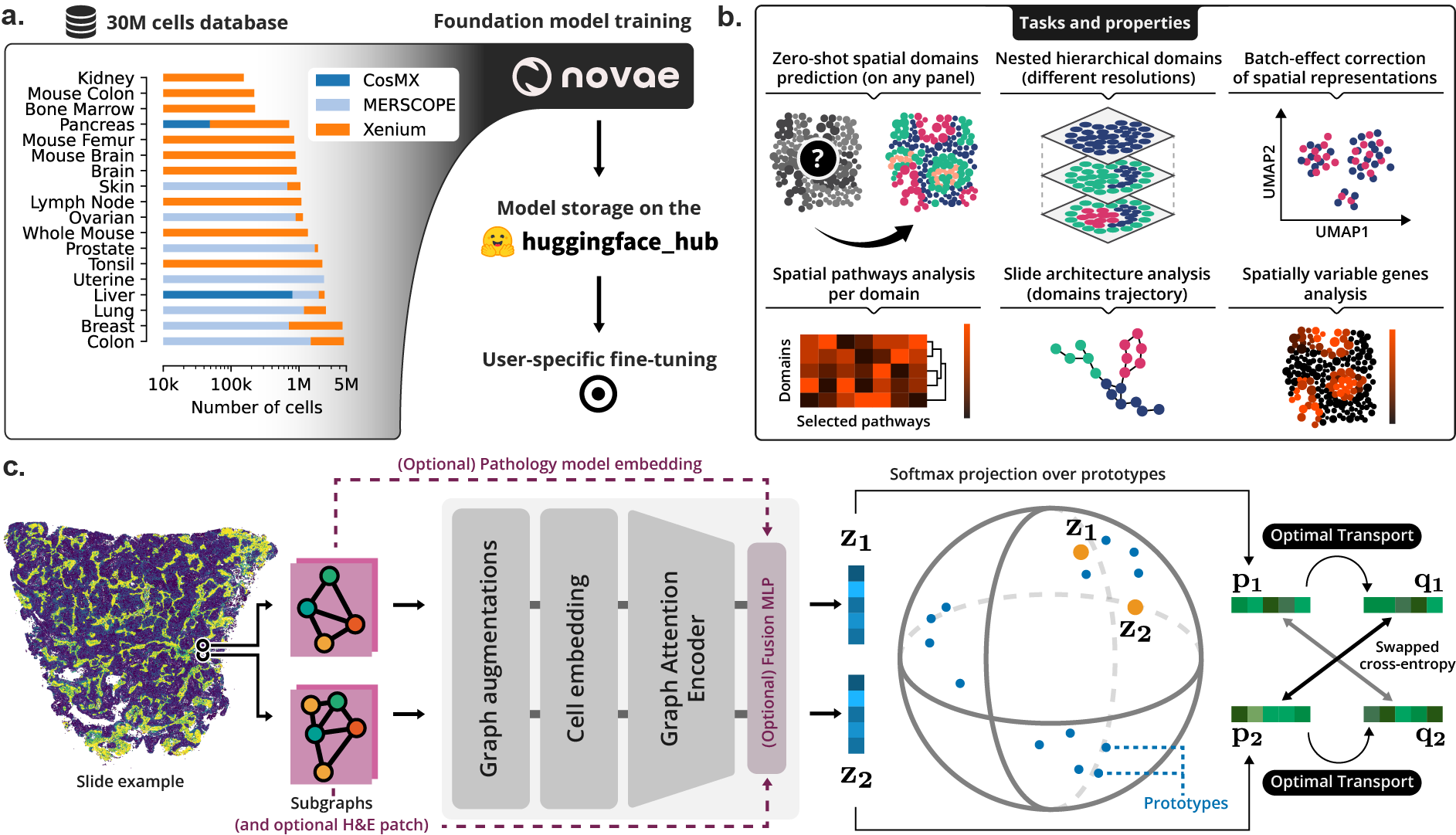
> **(a)** Novae was trained on a large dataset, and is shared on [Hugging Face Hub](https://huggingface.co/collections/MICS-Lab/novae-669cdf1754729d168a69f6bd). **(b)** Illustration of the main tasks and properties of Novae. **(c)** Illustration of the method behing Novae (self-supervision on graphs, adapted from [SwAV](https://arxiv.org/abs/2006.09882)).
diff --git a/docs/index.md b/docs/index.md
index b1b6b30..ef0eb72 100644
--- a/docs/index.md
+++ b/docs/index.md
@@ -2,4 +2,25 @@

-TODO
+
+ 💫 Graph-based foundation model for spatial transcriptomics data
+
+
+Novae is a deep learning model for spatial domain assignments of spatial transcriptomics data (at both single-cell or spot resolution). It works across multiple gene panels, tissues, and technologies. Novae offers several additional features, including: (i) native batch-effect correction, (ii) analysis of spatially variable genes and pathways, and (iii) architecture analysis of tissue slides.
+
+## Overview
+
+
+  +
+
+
+> **(a)** Novae was trained on a large dataset, and is shared on [Hugging Face Hub](https://huggingface.co/collections/MICS-Lab/novae-669cdf1754729d168a69f6bd). **(b)** Illustration of the main tasks and properties of Novae. **(c)** Illustration of the method behing Novae (self-supervision on graphs, adapted from [SwAV](https://arxiv.org/abs/2006.09882)).
+
+
+## Why using Novae
+
+- It is already pretrained on a large dataset (pan human/mouse tissues, brain, ...). Therefore, you can compute spatial domains in a zero-shot manner (i.e., without fine-tuning).
+- It has been developed to find consistent domain across many slides. This also work if you have different technologies (e.g., MERSCOPE/Xenium) and multiple gene panels.
+- You can natively correct batch effect, without using external tools.
+- After inference, the spatial domain assignment is super fast, allowing to try multiple resolutions easily.
+- It supports many downstream tasks, all included inside one framework.
diff --git a/pyproject.toml b/pyproject.toml
index 60f1adf..ab170bb 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -1,6 +1,6 @@
[tool.poetry]
name = "novae"
-version = "0.0.2"
+version = "0.0.5"
description = "Graph-based foundation model for spatial transcriptomics data"
documentation = "https://mics-lab.github.io/novae/"
homepage = "https://mics-lab.github.io/novae/"
 +
+ 
 +
+ 
 +
+ 
 -TODO
+
-TODO
+ +
+