MBfMRI is a unified Python fMRI analysis tool on task-based fMRI data to investigate brain implementations of latent neurocognitive processes. It combines two fMRI analytic frameworks: model-based fMRI and multi-voxel pattern analysis (MVPA). MBfMRI offers simple executable functions to conduct computational modeling (supported by hBayesDM[1]) and run model-based fMRI analysis using MVPA. To install.
The basic framework of conventional model-based fMRI by O'Doherty et al. (2007)[2] consists of the following steps.
- Find best fitting parameters of model to behavioral data
- Generate model-based time series by using the best model-fitting parameters and convolve it with HRF (a.k.a. latent process)
- Regress model-based time series against task-fMRI data using GLM
Upon the more conventional massive univariate approach to model-based neuroimaging, MBfMRI extends the framework by adopting MVPA regression models. The MVPA approach (model-based MVPA) has two differences compared to the previous approach: first, MVPA regression models predict cognitive process directly from brain activations, enabling acquisition of reverse inference model denoted by Poldrack (2006)[3]; second, instead of being mapped by statistical significance, the brain activation pattern correlated with the latent process is obtained by interpreting trained MVPA regression models.
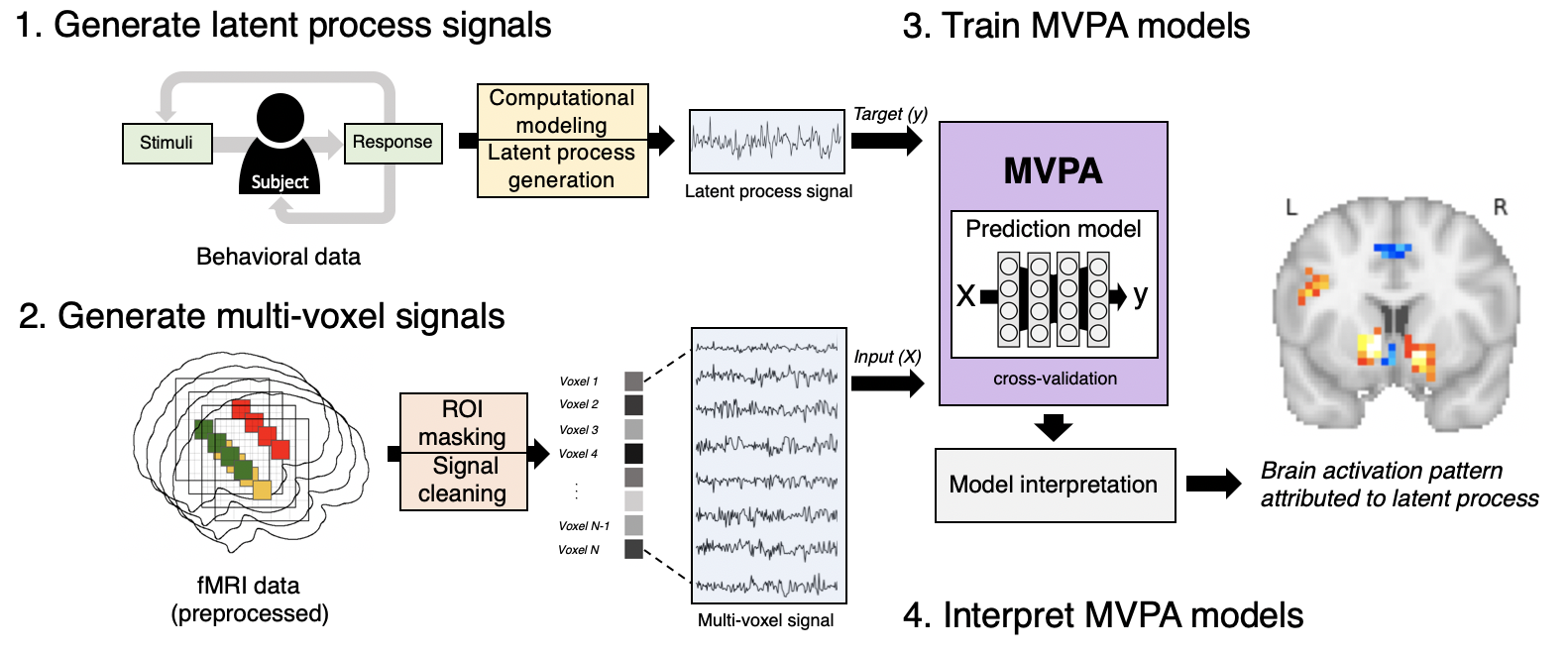
The specific workflow of the model-based MVPA, consists of the following steps:
- Generate latent process signals by fitting computational models with behavioral data and convolving time series of latent process with HRF.
- Generate multi-voxel signals from preprocess fMRI images by allowing ROI masking, zooming spatial resolution, and improving the quality of signals by several well-established methods (e.g. detrending, high-pass filtering, regressing out confounds).
- Train MVPA models by feeding multi-voxel signals as input (X) and latent process signals as ouput (y), or target, employing the repeated cross-validation framework.
- Interpret the trained MVPA models to visualize the brain implementation of the target latent process quantified as brain activation pattern attributed to predict the target signals from the multi-voxel signals.
Other distinguishing features of the model-based MVPA are that the model-based MVPA is flexible as it allows various MVPA models plugged in and it is free of analytic hierarchy (e.g. first-level anal. or second-level anal.).
The package also provides the well-established model-based GLM approach. The process shared with the model-based MVPA approach is 1) Generate latent process signals to provide parametric modulation of the target signals. The first-level and second-level analysis are done by NiLearn modules, FirstLevelModel and SecondLevelModel respectively. Please refer to the links if you'd like to know the details about each step.
MBfMRI supports Python 3.6 or above and relies on NiLearn, hBayesDM, py-glmnet, and tensorflow(tested on v2.4.0).
- Model-based MVPA is based on MVPA regression model.
- Model-based MVPA is flexible as it allows various MVPA models plugged in.
- Model-based MVPA is free of analytic hierarchy (e.g. first-level anal. or second-level anal.).
- MBfMRI offers various MVPA models as well as a massive univariate approach based on GLM.
git clone https://github.com/CCS-Lab/project_model_based_fmri.git
cd project_model_based_fmri
poetry install
poetry shell
python setup.py install
Download the example data from here. Download and unzip it under tutorials.
See the tutorial.
[1] Ahn, W.-Y., Haines, N., & Zhang, L. (2017). Revealing Neurocomputational Mechanisms of Reinforcement Learning and Decision-Making With the hBayesDM Package. Computational Psychiatry, 1(Figure 1), 24–57. https://doi.org/10.1162/cpsy_a_00002
[2] O’Doherty, J. P., Hampton, A., & Kim, H. (2007). Model-based fMRI and its application to reward learning and decision making. Annals of the New York Academy of Sciences, 1104, 35–53. https://doi.org/10.1196/annals.1390.022
[3] Poldrack, R. A. (2006). Can cognitive processes be inferred from neuroimaging data? Trends in Cognitive Sciences, 10(2), 59–63. https://doi.org/10.1016/j.tics.2005.12.004